Magnesium »
PDB 7pjf-7pn8 »
7pm9 »
Magnesium in PDB 7pm9: Cryo-Em Structure of the Actomyosin-V Complex in the Strong-Adp State (Central 1ER, Class 4)
Magnesium Binding Sites:
The binding sites of Magnesium atom in the Cryo-Em Structure of the Actomyosin-V Complex in the Strong-Adp State (Central 1ER, Class 4)
(pdb code 7pm9). This binding sites where shown within
5.0 Angstroms radius around Magnesium atom.
In total 2 binding sites of Magnesium where determined in the Cryo-Em Structure of the Actomyosin-V Complex in the Strong-Adp State (Central 1ER, Class 4), PDB code: 7pm9:
Jump to Magnesium binding site number: 1; 2;
In total 2 binding sites of Magnesium where determined in the Cryo-Em Structure of the Actomyosin-V Complex in the Strong-Adp State (Central 1ER, Class 4), PDB code: 7pm9:
Jump to Magnesium binding site number: 1; 2;
Magnesium binding site 1 out of 2 in 7pm9
Go back to
Magnesium binding site 1 out
of 2 in the Cryo-Em Structure of the Actomyosin-V Complex in the Strong-Adp State (Central 1ER, Class 4)
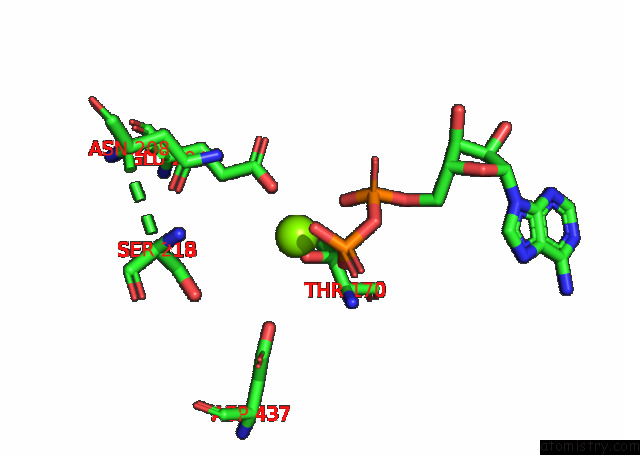
Mono view
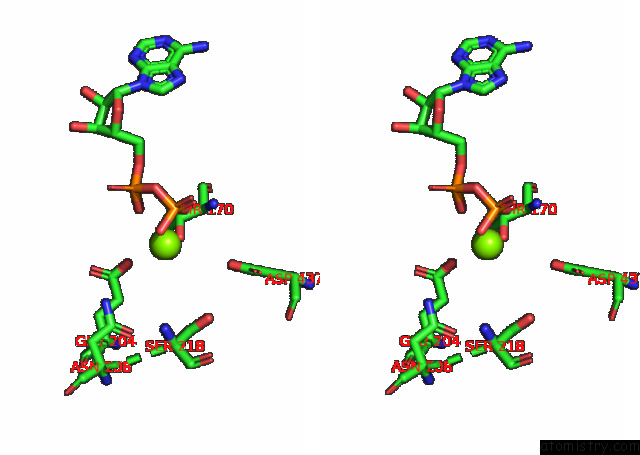
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 1 of Cryo-Em Structure of the Actomyosin-V Complex in the Strong-Adp State (Central 1ER, Class 4) within 5.0Å range:
|
Magnesium binding site 2 out of 2 in 7pm9
Go back to
Magnesium binding site 2 out
of 2 in the Cryo-Em Structure of the Actomyosin-V Complex in the Strong-Adp State (Central 1ER, Class 4)
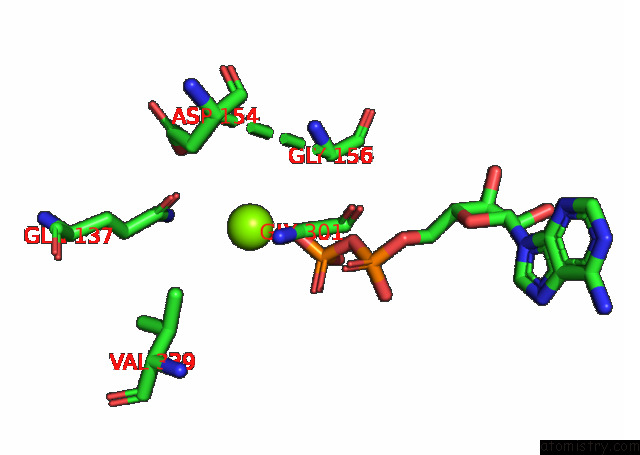
Mono view
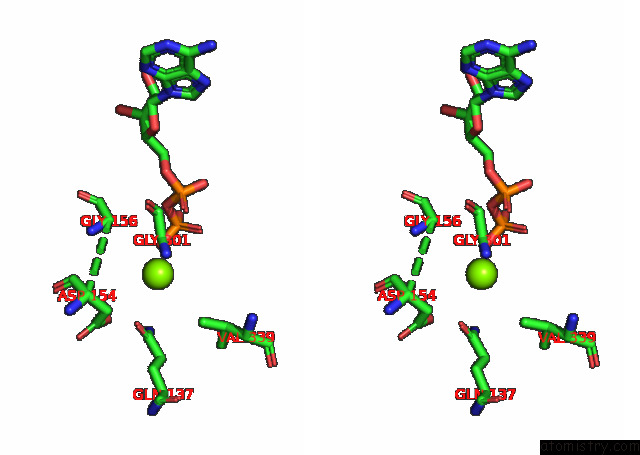
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 2 of Cryo-Em Structure of the Actomyosin-V Complex in the Strong-Adp State (Central 1ER, Class 4) within 5.0Å range:
|
Reference:
S.Pospich,
H.L.Sweeney,
A.Houdusse,
S.Raunser.
High-Resolution Structures of the Actomyosin-V Complex in Three Nucleotide States Provide Insights Into the Force Generation Mechanism. Elife V. 10 2021.
ISSN: ESSN 2050-084X
PubMed: 34812732
DOI: 10.7554/ELIFE.73724
Page generated: Thu Oct 3 04:40:08 2024
ISSN: ESSN 2050-084X
PubMed: 34812732
DOI: 10.7554/ELIFE.73724
Last articles
Ca in 5UE1Ca in 5UE2
Ca in 5UCQ
Ca in 5UDL
Ca in 5UDK
Ca in 5UD6
Ca in 5UDJ
Ca in 5UDI
Ca in 5UD0
Ca in 5UBJ