Magnesium »
PDB 7r8p-7rf0 »
7r8q »
Magnesium in PDB 7r8q: Closed Form of SAOUHSC_02373 in Complex with Adp, Citrate, MG2+ and Na+
Protein crystallography data
The structure of Closed Form of SAOUHSC_02373 in Complex with Adp, Citrate, MG2+ and Na+, PDB code: 7r8q
was solved by
J.L.Pederick,
J.B.Bruning,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 36.89 / 2.00 |
| Space group | C 1 2 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 145.868, 82.812, 107.782, 90, 131.06, 90 |
| R / Rfree (%) | 19.1 / 22 |
Other elements in 7r8q:
The structure of Closed Form of SAOUHSC_02373 in Complex with Adp, Citrate, MG2+ and Na+ also contains other interesting chemical elements:
| Sodium | (Na) | 6 atoms |
Magnesium Binding Sites:
The binding sites of Magnesium atom in the Closed Form of SAOUHSC_02373 in Complex with Adp, Citrate, MG2+ and Na+
(pdb code 7r8q). This binding sites where shown within
5.0 Angstroms radius around Magnesium atom.
In total 2 binding sites of Magnesium where determined in the Closed Form of SAOUHSC_02373 in Complex with Adp, Citrate, MG2+ and Na+, PDB code: 7r8q:
Jump to Magnesium binding site number: 1; 2;
In total 2 binding sites of Magnesium where determined in the Closed Form of SAOUHSC_02373 in Complex with Adp, Citrate, MG2+ and Na+, PDB code: 7r8q:
Jump to Magnesium binding site number: 1; 2;
Magnesium binding site 1 out of 2 in 7r8q
Go back to
Magnesium binding site 1 out
of 2 in the Closed Form of SAOUHSC_02373 in Complex with Adp, Citrate, MG2+ and Na+
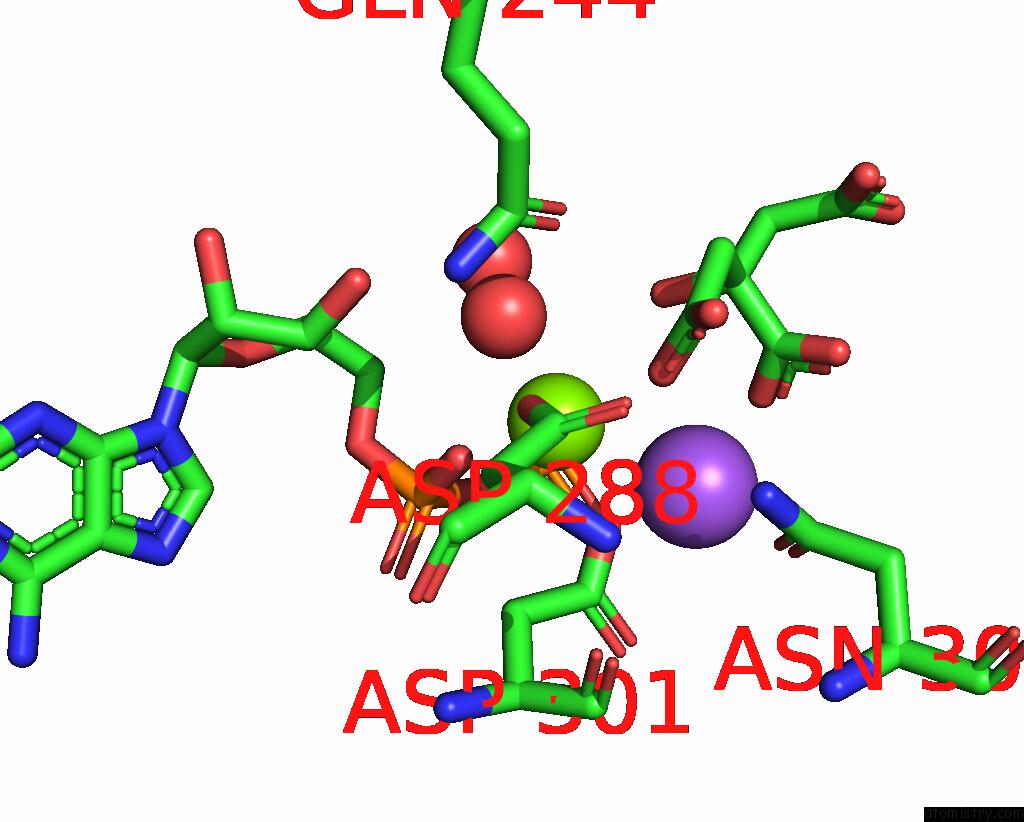
Mono view
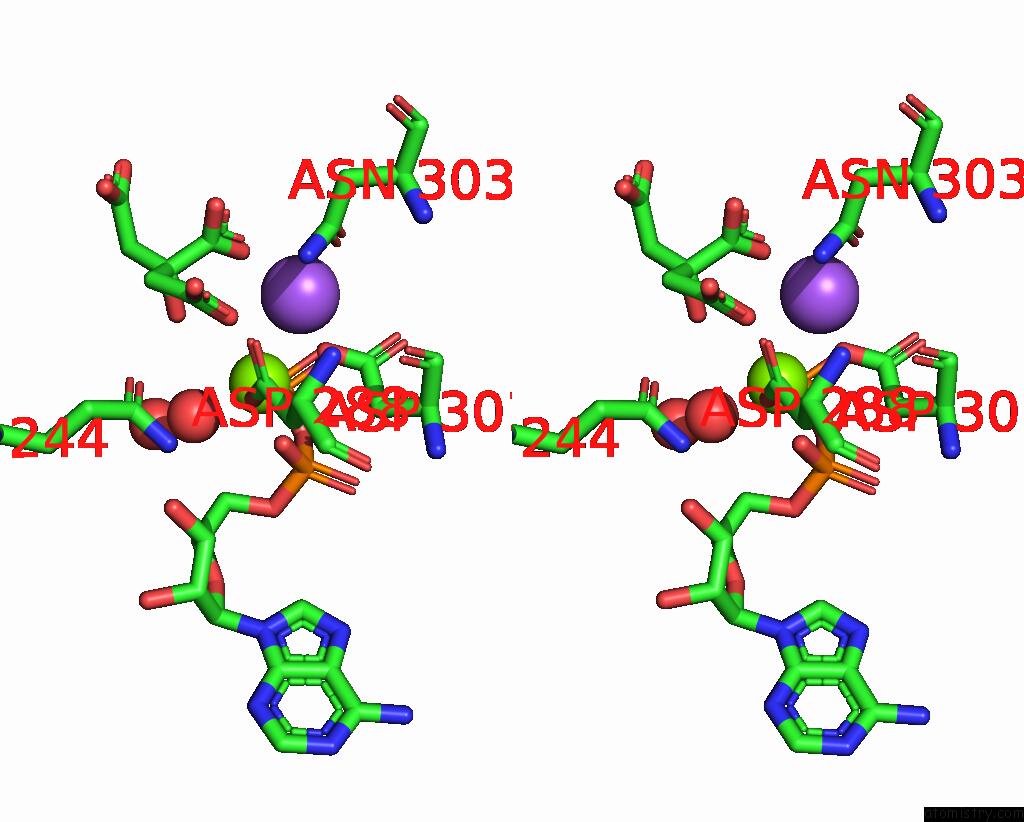
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 1 of Closed Form of SAOUHSC_02373 in Complex with Adp, Citrate, MG2+ and Na+ within 5.0Å range:
|
Magnesium binding site 2 out of 2 in 7r8q
Go back to
Magnesium binding site 2 out
of 2 in the Closed Form of SAOUHSC_02373 in Complex with Adp, Citrate, MG2+ and Na+
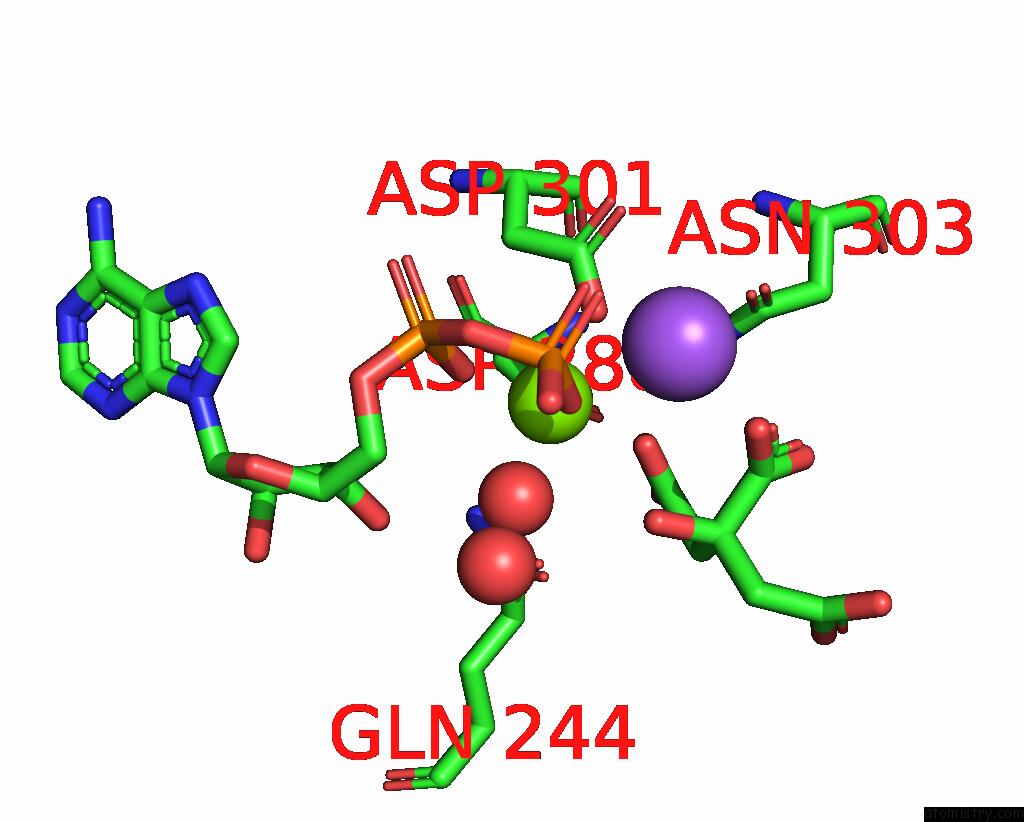
Mono view
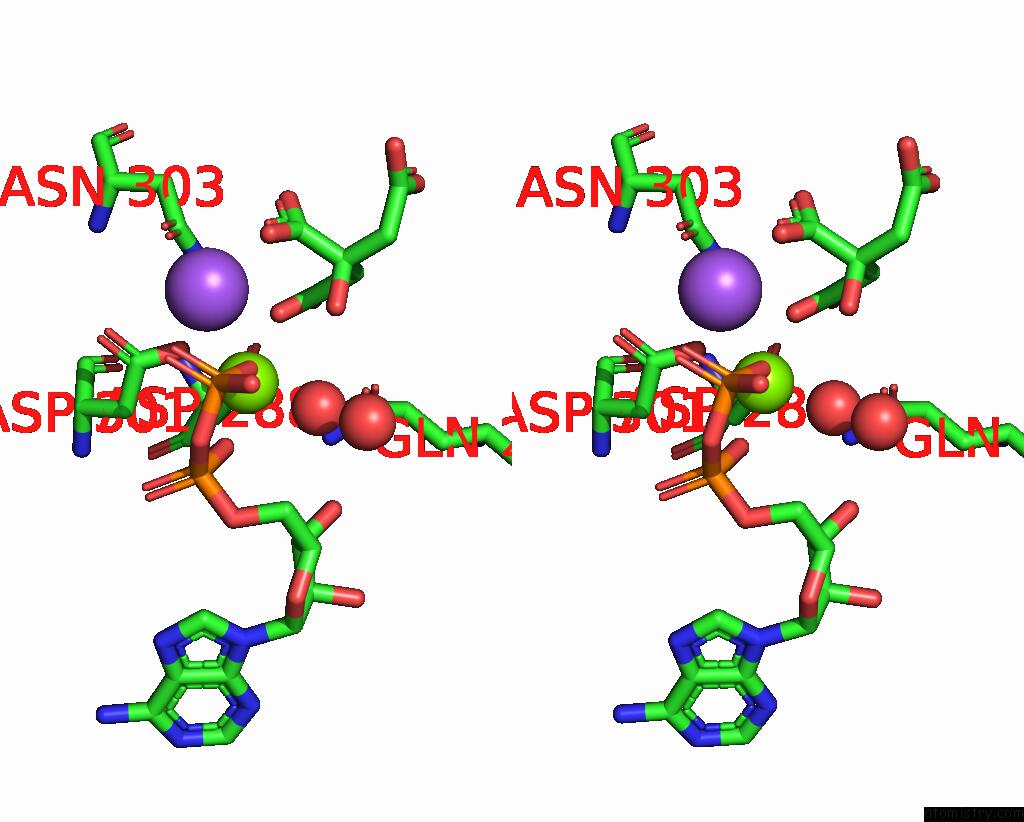
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 2 of Closed Form of SAOUHSC_02373 in Complex with Adp, Citrate, MG2+ and Na+ within 5.0Å range:
|
Reference:
J.L.Pederick,
A.J.Horsfall,
B.Jovcevski,
J.Klose,
A.D.Abell,
T.L.Pukala,
J.B.Bruning.
Discovery of An ʟ-Amino Acid Ligase Implicated in Staphylococcal Sulfur Amino Acid Metabolism. J.Biol.Chem. V. 298 02392 2022.
ISSN: ESSN 1083-351X
PubMed: 35988643
DOI: 10.1016/J.JBC.2022.102392
Page generated: Thu Aug 14 14:54:25 2025
ISSN: ESSN 1083-351X
PubMed: 35988643
DOI: 10.1016/J.JBC.2022.102392
Last articles
Mg in 7VZ1Mg in 7VYN
Mg in 7VYF
Mg in 7VYH
Mg in 7VY8
Mg in 7VXU
Mg in 7VYA
Mg in 7VRJ
Mg in 7VXQ
Mg in 7VXP