Magnesium »
PDB 7tf9-7tlk »
7tid »
Magnesium in PDB 7tid: Structure of the Yeast Clamp Loader (Replication Factor C Rfc) Bound to the Sliding Clamp (Proliferating Cell Nuclear Antigen Pcna) and Primer-Template Dna
Magnesium Binding Sites:
The binding sites of Magnesium atom in the Structure of the Yeast Clamp Loader (Replication Factor C Rfc) Bound to the Sliding Clamp (Proliferating Cell Nuclear Antigen Pcna) and Primer-Template Dna
(pdb code 7tid). This binding sites where shown within
5.0 Angstroms radius around Magnesium atom.
In total 4 binding sites of Magnesium where determined in the Structure of the Yeast Clamp Loader (Replication Factor C Rfc) Bound to the Sliding Clamp (Proliferating Cell Nuclear Antigen Pcna) and Primer-Template Dna, PDB code: 7tid:
Jump to Magnesium binding site number: 1; 2; 3; 4;
In total 4 binding sites of Magnesium where determined in the Structure of the Yeast Clamp Loader (Replication Factor C Rfc) Bound to the Sliding Clamp (Proliferating Cell Nuclear Antigen Pcna) and Primer-Template Dna, PDB code: 7tid:
Jump to Magnesium binding site number: 1; 2; 3; 4;
Magnesium binding site 1 out of 4 in 7tid
Go back to
Magnesium binding site 1 out
of 4 in the Structure of the Yeast Clamp Loader (Replication Factor C Rfc) Bound to the Sliding Clamp (Proliferating Cell Nuclear Antigen Pcna) and Primer-Template Dna

Mono view
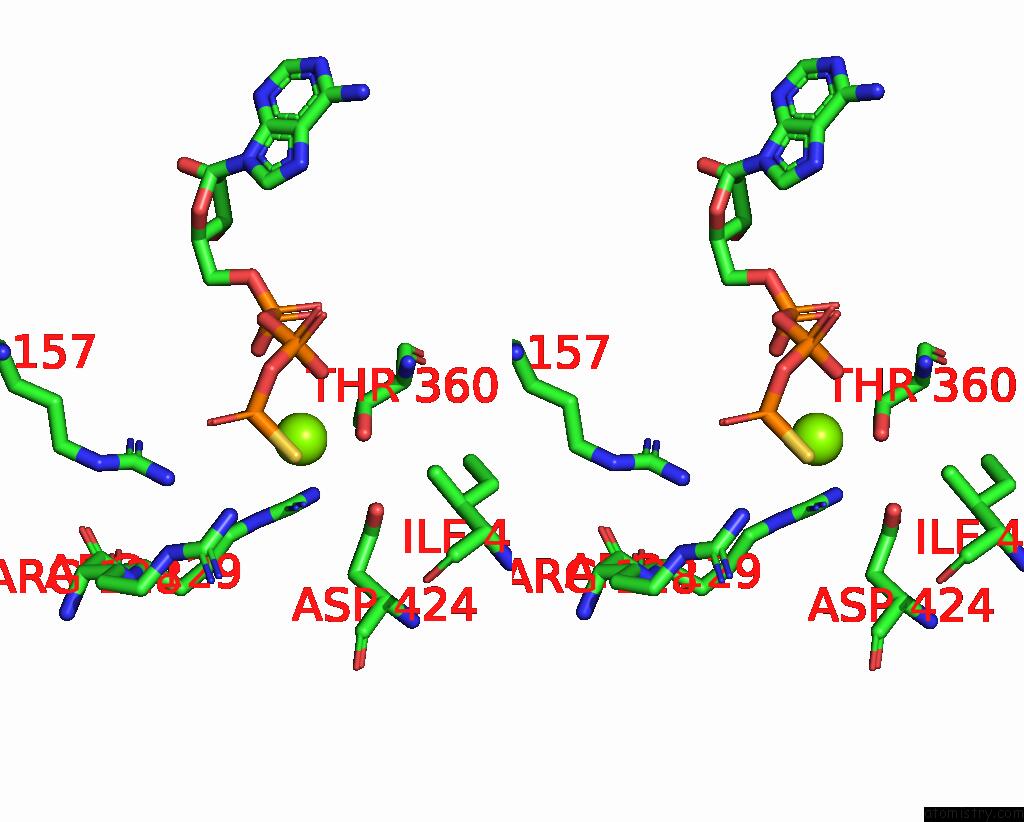
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 1 of Structure of the Yeast Clamp Loader (Replication Factor C Rfc) Bound to the Sliding Clamp (Proliferating Cell Nuclear Antigen Pcna) and Primer-Template Dna within 5.0Å range:
|
Magnesium binding site 2 out of 4 in 7tid
Go back to
Magnesium binding site 2 out
of 4 in the Structure of the Yeast Clamp Loader (Replication Factor C Rfc) Bound to the Sliding Clamp (Proliferating Cell Nuclear Antigen Pcna) and Primer-Template Dna
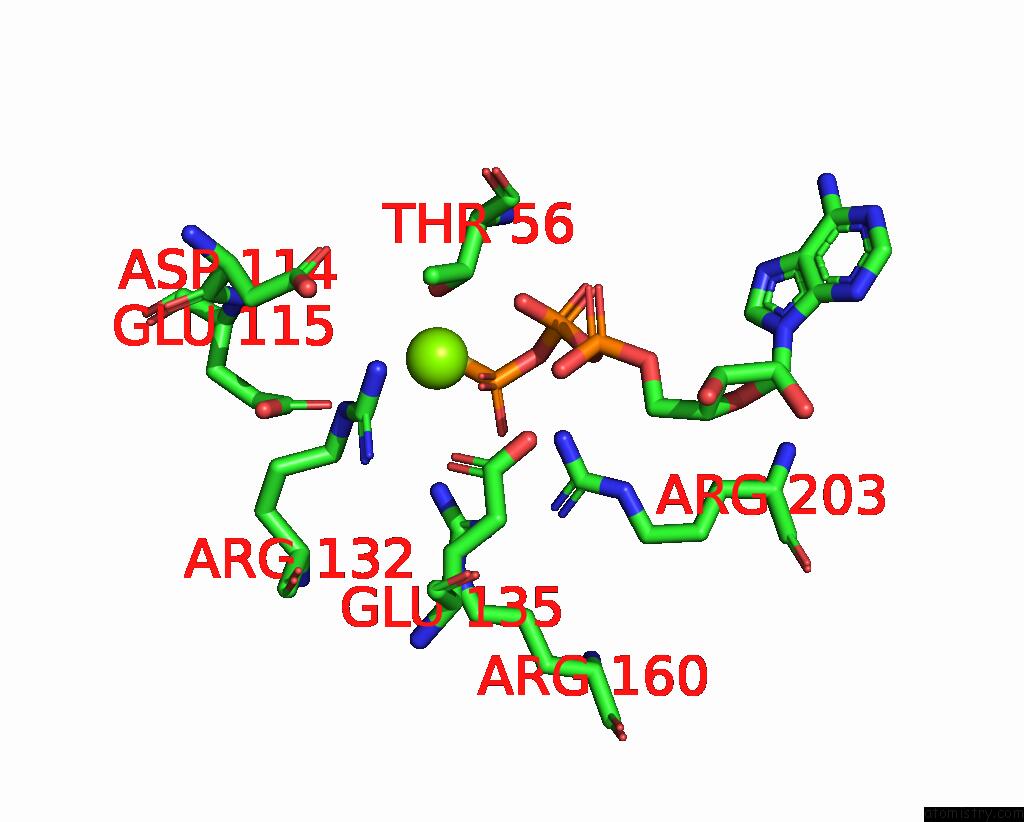
Mono view
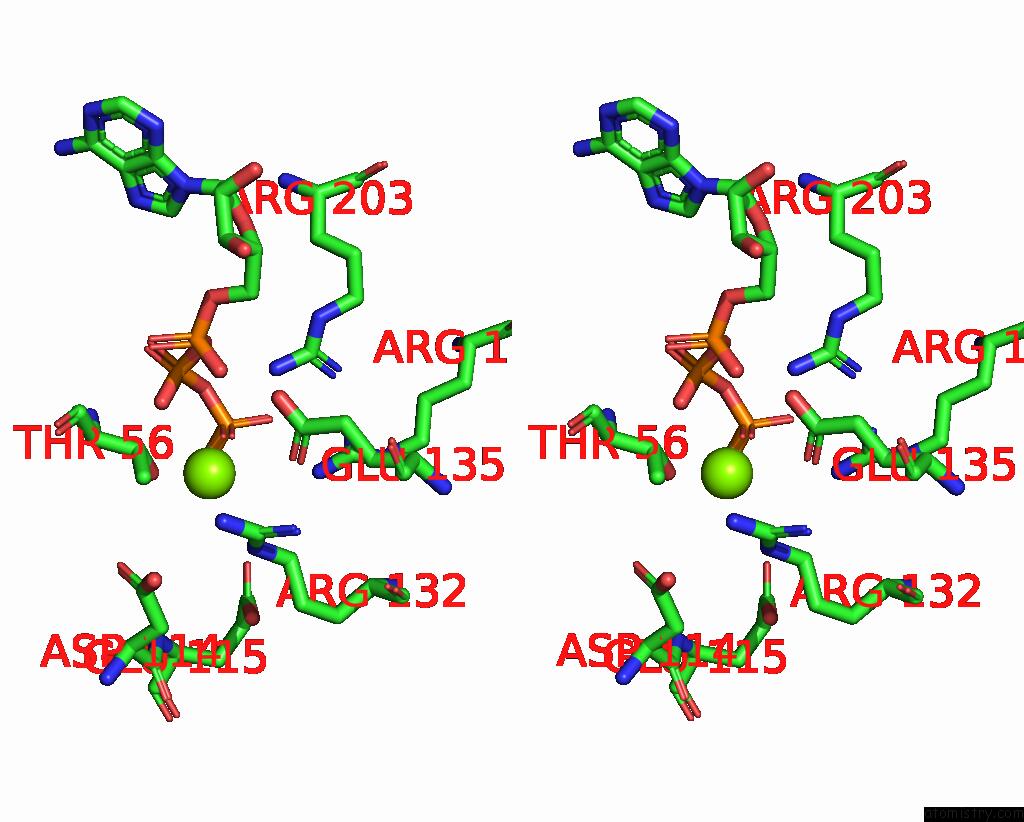
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 2 of Structure of the Yeast Clamp Loader (Replication Factor C Rfc) Bound to the Sliding Clamp (Proliferating Cell Nuclear Antigen Pcna) and Primer-Template Dna within 5.0Å range:
|
Magnesium binding site 3 out of 4 in 7tid
Go back to
Magnesium binding site 3 out
of 4 in the Structure of the Yeast Clamp Loader (Replication Factor C Rfc) Bound to the Sliding Clamp (Proliferating Cell Nuclear Antigen Pcna) and Primer-Template Dna

Mono view
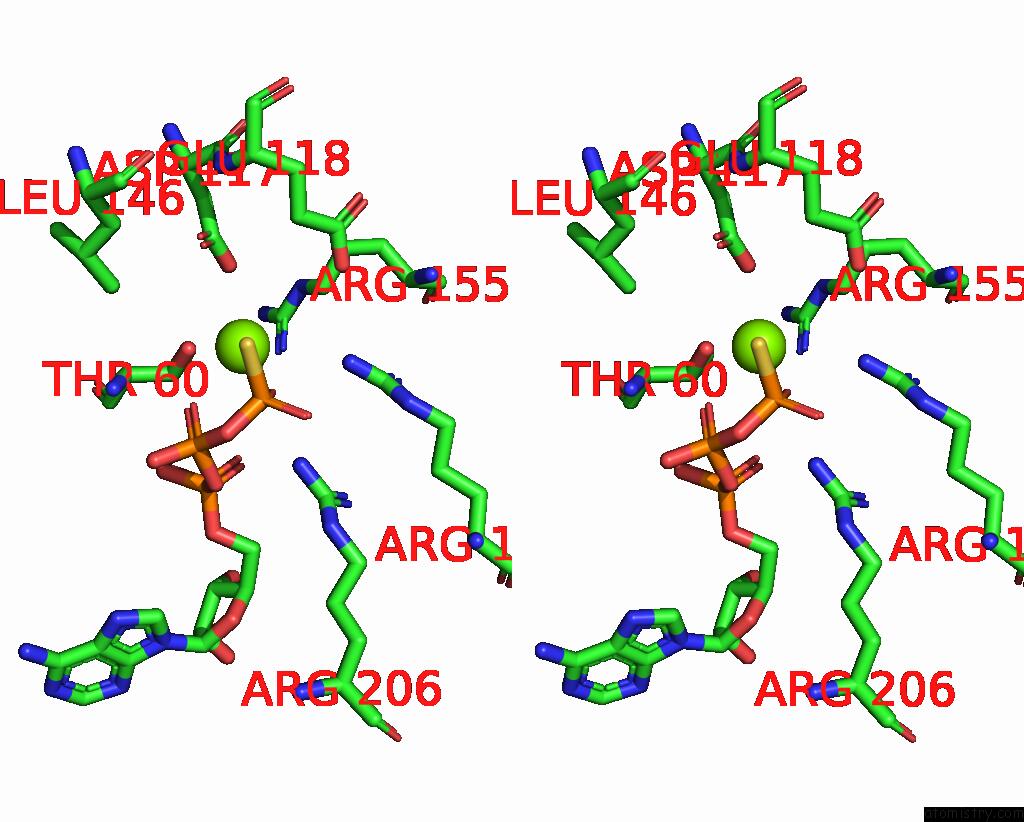
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 3 of Structure of the Yeast Clamp Loader (Replication Factor C Rfc) Bound to the Sliding Clamp (Proliferating Cell Nuclear Antigen Pcna) and Primer-Template Dna within 5.0Å range:
|
Magnesium binding site 4 out of 4 in 7tid
Go back to
Magnesium binding site 4 out
of 4 in the Structure of the Yeast Clamp Loader (Replication Factor C Rfc) Bound to the Sliding Clamp (Proliferating Cell Nuclear Antigen Pcna) and Primer-Template Dna
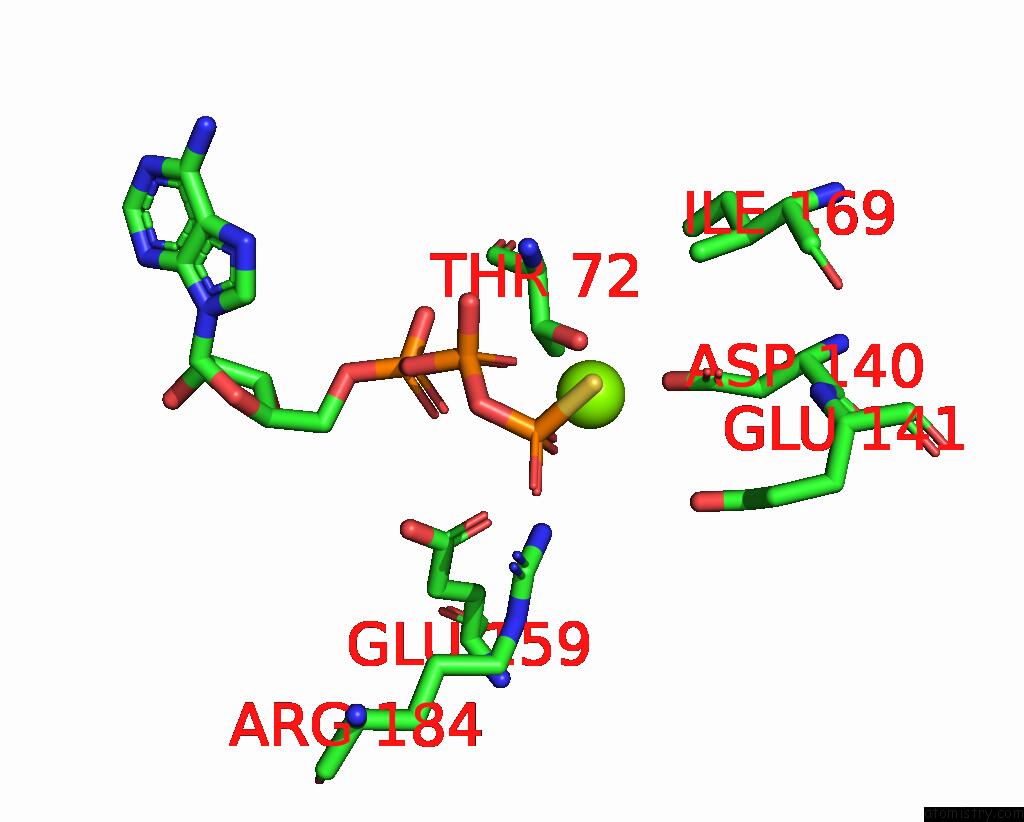
Mono view
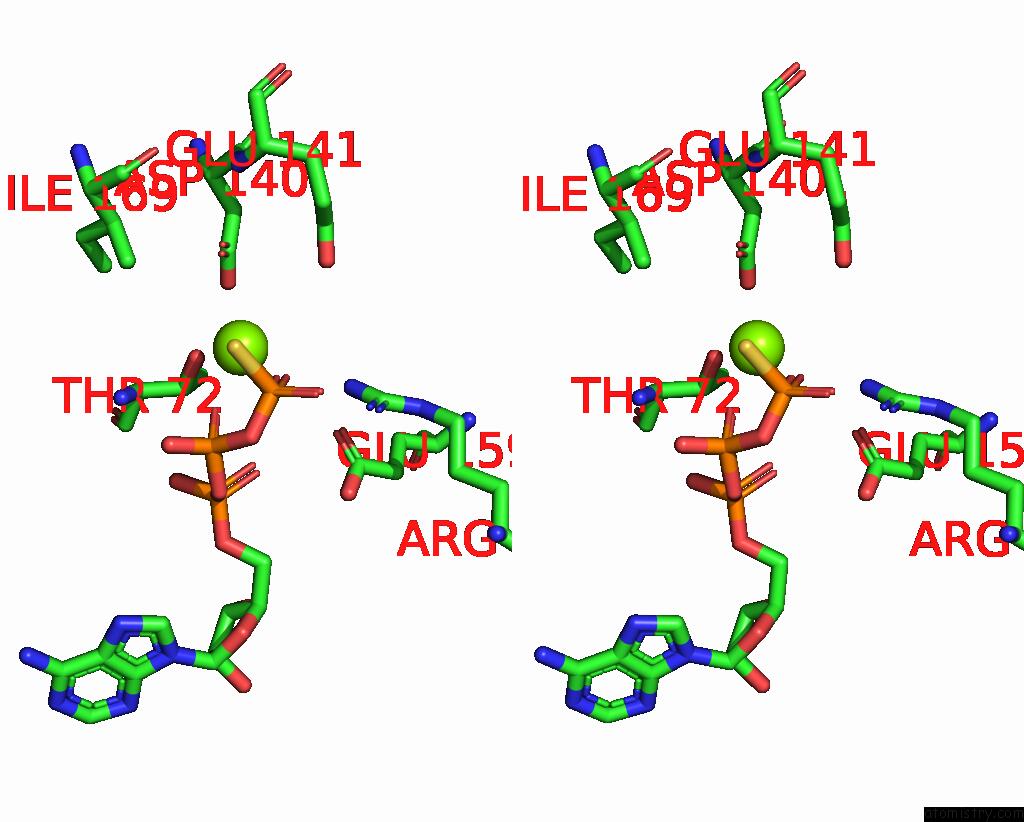
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 4 of Structure of the Yeast Clamp Loader (Replication Factor C Rfc) Bound to the Sliding Clamp (Proliferating Cell Nuclear Antigen Pcna) and Primer-Template Dna within 5.0Å range:
|
Reference:
C.Gaubitz,
X.Liu,
J.Pajak,
N.P.Stone,
J.A.Hayes,
G.Demo,
B.A.Kelch Phd.
Cryo-Em Structures Reveal High-Resolution Mechanism of A Dna Polymerase Sliding Clamp Loader. Elife V. 11 2022.
ISSN: ESSN 2050-084X
PubMed: 35179493
DOI: 10.7554/ELIFE.74175
Page generated: Thu Oct 3 09:16:45 2024
ISSN: ESSN 2050-084X
PubMed: 35179493
DOI: 10.7554/ELIFE.74175
Last articles
Cl in 6BV8Cl in 6BV5
Cl in 6BV9
Cl in 6BUQ
Cl in 6BUP
Cl in 6BTW
Cl in 6BUN
Cl in 6BU6
Cl in 6BUM
Cl in 6BSQ