Magnesium »
PDB 7ywa-7z2c »
7z16 »
Magnesium in PDB 7z16: E. Coli C-P Lyase Bound to Phnk/Phnl Dual Abc Dimer with Amppnp and Phnk E171Q Mutation
Enzymatic activity of E. Coli C-P Lyase Bound to Phnk/Phnl Dual Abc Dimer with Amppnp and Phnk E171Q Mutation
All present enzymatic activity of E. Coli C-P Lyase Bound to Phnk/Phnl Dual Abc Dimer with Amppnp and Phnk E171Q Mutation:
2.7.8.37; 4.7.1.1;
2.7.8.37; 4.7.1.1;
Other elements in 7z16:
The structure of E. Coli C-P Lyase Bound to Phnk/Phnl Dual Abc Dimer with Amppnp and Phnk E171Q Mutation also contains other interesting chemical elements:
| Zinc | (Zn) | 4 atoms |
Magnesium Binding Sites:
The binding sites of Magnesium atom in the E. Coli C-P Lyase Bound to Phnk/Phnl Dual Abc Dimer with Amppnp and Phnk E171Q Mutation
(pdb code 7z16). This binding sites where shown within
5.0 Angstroms radius around Magnesium atom.
In total 2 binding sites of Magnesium where determined in the E. Coli C-P Lyase Bound to Phnk/Phnl Dual Abc Dimer with Amppnp and Phnk E171Q Mutation, PDB code: 7z16:
Jump to Magnesium binding site number: 1; 2;
In total 2 binding sites of Magnesium where determined in the E. Coli C-P Lyase Bound to Phnk/Phnl Dual Abc Dimer with Amppnp and Phnk E171Q Mutation, PDB code: 7z16:
Jump to Magnesium binding site number: 1; 2;
Magnesium binding site 1 out of 2 in 7z16
Go back to
Magnesium binding site 1 out
of 2 in the E. Coli C-P Lyase Bound to Phnk/Phnl Dual Abc Dimer with Amppnp and Phnk E171Q Mutation
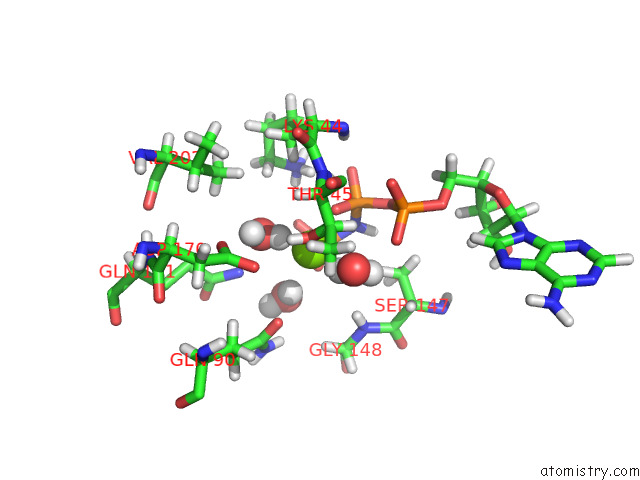
Mono view
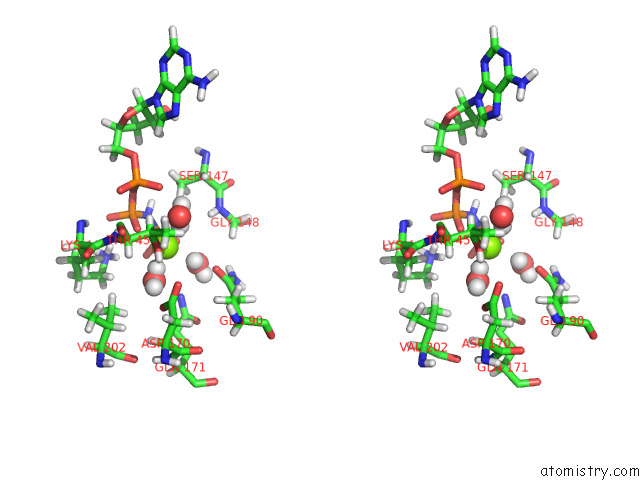
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 1 of E. Coli C-P Lyase Bound to Phnk/Phnl Dual Abc Dimer with Amppnp and Phnk E171Q Mutation within 5.0Å range:
|
Magnesium binding site 2 out of 2 in 7z16
Go back to
Magnesium binding site 2 out
of 2 in the E. Coli C-P Lyase Bound to Phnk/Phnl Dual Abc Dimer with Amppnp and Phnk E171Q Mutation
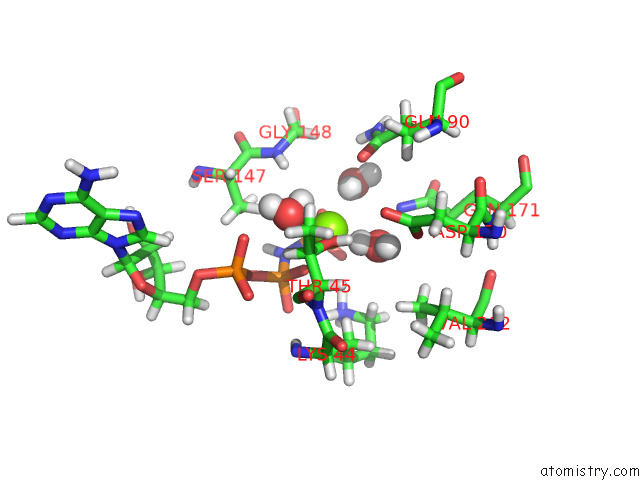
Mono view
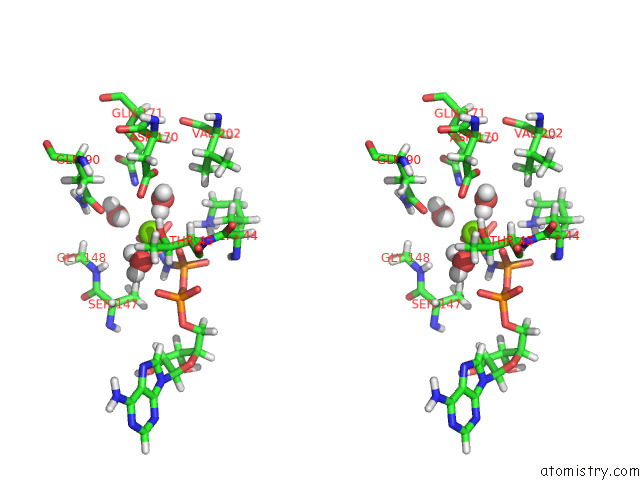
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 2 of E. Coli C-P Lyase Bound to Phnk/Phnl Dual Abc Dimer with Amppnp and Phnk E171Q Mutation within 5.0Å range:
|
Reference:
S.K.Amstrup,
N.Sofos,
J.L.Karlsen,
R.B.Skjerning,
T.Boesen,
J.J.Enghild,
B.Hove-Jensen,
D.E.Brodersen.
Structural Remodelling of the Carbon-Phosphorus Lyase Machinery By A Dual Abc Atpase Biorxiv 2022.
DOI: 10.1101/2022.06.09.495270
Page generated: Thu Oct 3 16:13:36 2024
DOI: 10.1101/2022.06.09.495270
Last articles
Fe in 2YXOFe in 2YRS
Fe in 2YXC
Fe in 2YNM
Fe in 2YVJ
Fe in 2YP1
Fe in 2YU2
Fe in 2YU1
Fe in 2YQB
Fe in 2YOO