Magnesium »
PDB 7zmu-7zz2 »
7zxy »
Magnesium in PDB 7zxy: 3.15 Angstrom Cryo-Em Structure of the Dimeric Cytochrome B6F Complex From Synechocystis Sp. Pcc 6803 with Natively Bound Plastoquinone and Lipid Molecules.
Enzymatic activity of 3.15 Angstrom Cryo-Em Structure of the Dimeric Cytochrome B6F Complex From Synechocystis Sp. Pcc 6803 with Natively Bound Plastoquinone and Lipid Molecules.
All present enzymatic activity of 3.15 Angstrom Cryo-Em Structure of the Dimeric Cytochrome B6F Complex From Synechocystis Sp. Pcc 6803 with Natively Bound Plastoquinone and Lipid Molecules.:
7.1.1.6;
7.1.1.6;
Other elements in 7zxy:
The structure of 3.15 Angstrom Cryo-Em Structure of the Dimeric Cytochrome B6F Complex From Synechocystis Sp. Pcc 6803 with Natively Bound Plastoquinone and Lipid Molecules. also contains other interesting chemical elements:
| Iron | (Fe) | 12 atoms |
Magnesium Binding Sites:
The binding sites of Magnesium atom in the 3.15 Angstrom Cryo-Em Structure of the Dimeric Cytochrome B6F Complex From Synechocystis Sp. Pcc 6803 with Natively Bound Plastoquinone and Lipid Molecules.
(pdb code 7zxy). This binding sites where shown within
5.0 Angstroms radius around Magnesium atom.
In total 2 binding sites of Magnesium where determined in the 3.15 Angstrom Cryo-Em Structure of the Dimeric Cytochrome B6F Complex From Synechocystis Sp. Pcc 6803 with Natively Bound Plastoquinone and Lipid Molecules., PDB code: 7zxy:
Jump to Magnesium binding site number: 1; 2;
In total 2 binding sites of Magnesium where determined in the 3.15 Angstrom Cryo-Em Structure of the Dimeric Cytochrome B6F Complex From Synechocystis Sp. Pcc 6803 with Natively Bound Plastoquinone and Lipid Molecules., PDB code: 7zxy:
Jump to Magnesium binding site number: 1; 2;
Magnesium binding site 1 out of 2 in 7zxy
Go back to
Magnesium binding site 1 out
of 2 in the 3.15 Angstrom Cryo-Em Structure of the Dimeric Cytochrome B6F Complex From Synechocystis Sp. Pcc 6803 with Natively Bound Plastoquinone and Lipid Molecules.
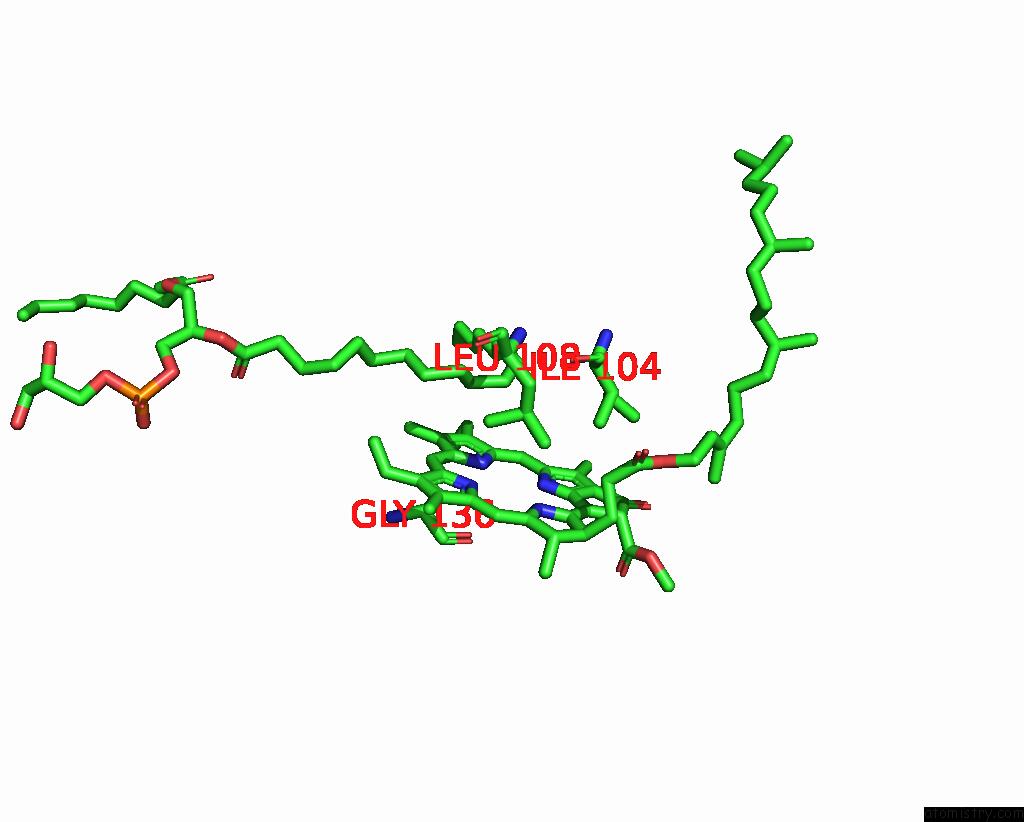
Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 1 of 3.15 Angstrom Cryo-Em Structure of the Dimeric Cytochrome B6F Complex From Synechocystis Sp. Pcc 6803 with Natively Bound Plastoquinone and Lipid Molecules. within 5.0Å range:
|
Magnesium binding site 2 out of 2 in 7zxy
Go back to
Magnesium binding site 2 out
of 2 in the 3.15 Angstrom Cryo-Em Structure of the Dimeric Cytochrome B6F Complex From Synechocystis Sp. Pcc 6803 with Natively Bound Plastoquinone and Lipid Molecules.
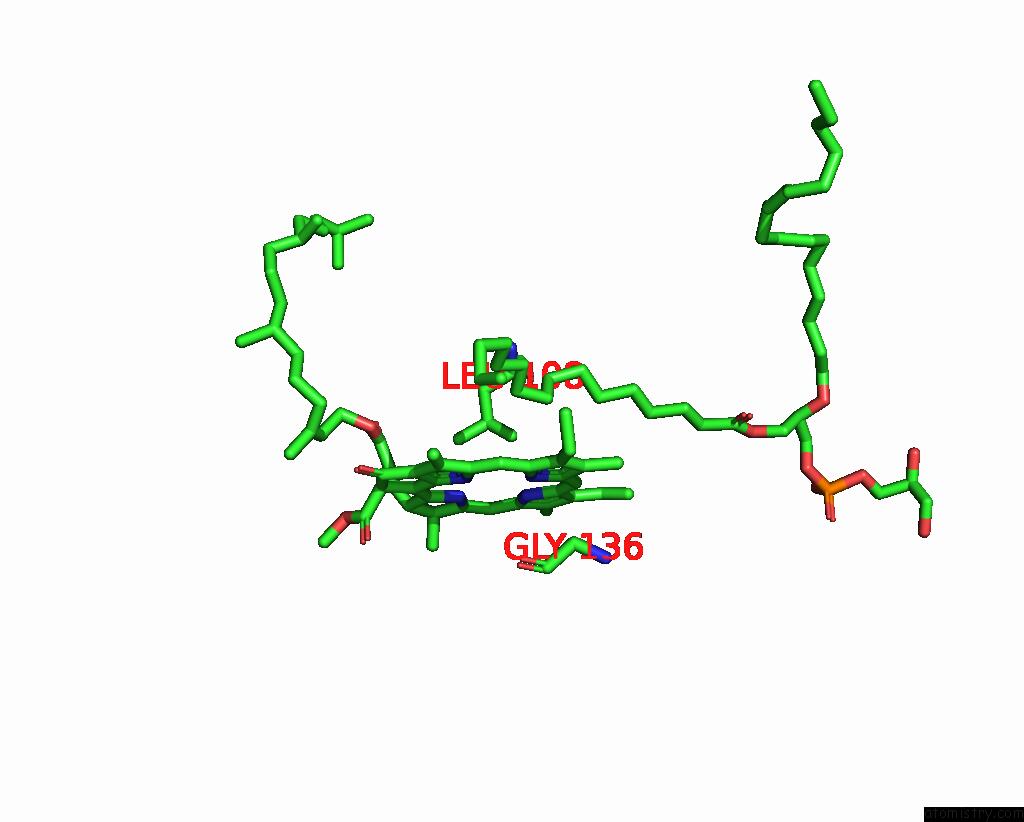
Mono view
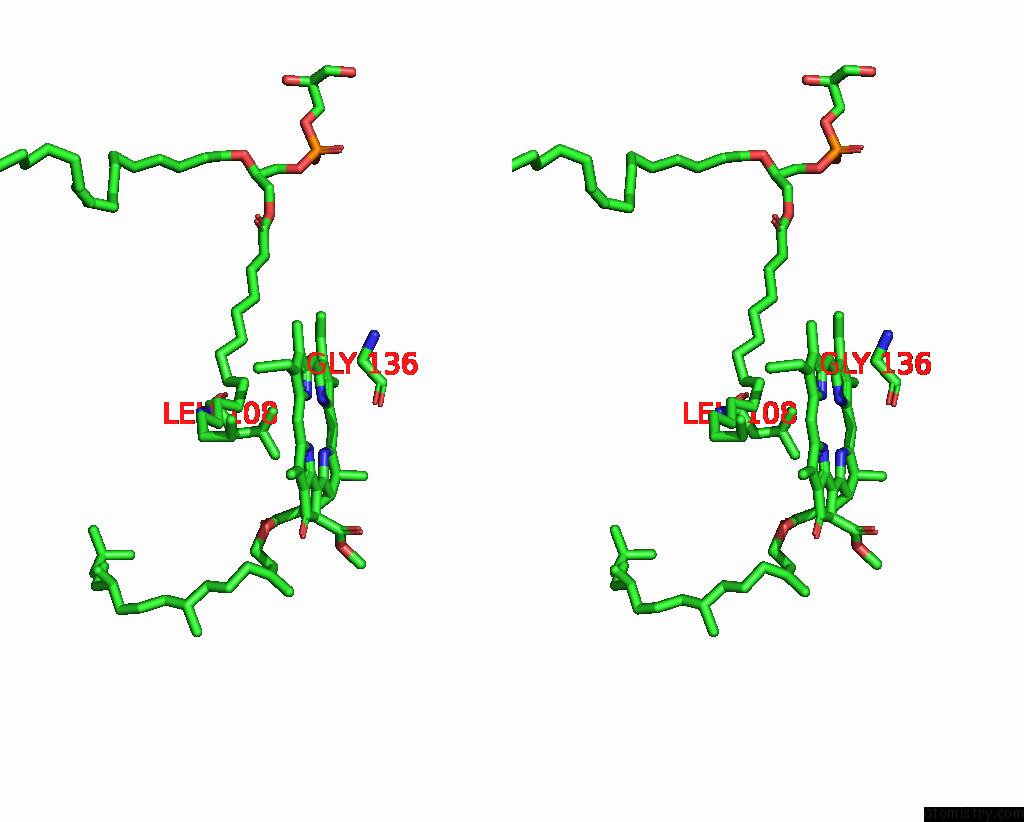
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 2 of 3.15 Angstrom Cryo-Em Structure of the Dimeric Cytochrome B6F Complex From Synechocystis Sp. Pcc 6803 with Natively Bound Plastoquinone and Lipid Molecules. within 5.0Å range:
|
Reference:
M.S.Proctor,
L.A.Malone,
D.A.Farmer,
D.J.K.Swainsbury,
F.R.Hawkings,
F.Pastorelli,
T.Z.Emrich-Mills,
C.A.Siebert,
C.N.Hunter,
M.P.Johnson,
A.Hitchcock.
Cryo-Em Structures of the Synechocystis Sp. Pcc 6803 Cytochrome B6F Complex with and Without the Regulatory Petp Subunit. Biochem.J. V. 479 1487 2022.
ISSN: ESSN 1470-8728
PubMed: 35726684
DOI: 10.1042/BCJ20220124
Page generated: Thu Oct 3 17:06:35 2024
ISSN: ESSN 1470-8728
PubMed: 35726684
DOI: 10.1042/BCJ20220124
Last articles
Fe in 2YXOFe in 2YRS
Fe in 2YXC
Fe in 2YNM
Fe in 2YVJ
Fe in 2YP1
Fe in 2YU2
Fe in 2YU1
Fe in 2YQB
Fe in 2YOO