Magnesium »
PDB 7zz3-8a63 »
8a5p »
Magnesium in PDB 8a5p: Structure of ARP4-IES4-N-Actin-ARP8-INO80HSA Subcomplex (A-Module) of Chaetomium Thermophilum INO80 on Curved Dna
Magnesium Binding Sites:
The binding sites of Magnesium atom in the Structure of ARP4-IES4-N-Actin-ARP8-INO80HSA Subcomplex (A-Module) of Chaetomium Thermophilum INO80 on Curved Dna
(pdb code 8a5p). This binding sites where shown within
5.0 Angstroms radius around Magnesium atom.
In total 3 binding sites of Magnesium where determined in the Structure of ARP4-IES4-N-Actin-ARP8-INO80HSA Subcomplex (A-Module) of Chaetomium Thermophilum INO80 on Curved Dna, PDB code: 8a5p:
Jump to Magnesium binding site number: 1; 2; 3;
In total 3 binding sites of Magnesium where determined in the Structure of ARP4-IES4-N-Actin-ARP8-INO80HSA Subcomplex (A-Module) of Chaetomium Thermophilum INO80 on Curved Dna, PDB code: 8a5p:
Jump to Magnesium binding site number: 1; 2; 3;
Magnesium binding site 1 out of 3 in 8a5p
Go back to
Magnesium binding site 1 out
of 3 in the Structure of ARP4-IES4-N-Actin-ARP8-INO80HSA Subcomplex (A-Module) of Chaetomium Thermophilum INO80 on Curved Dna
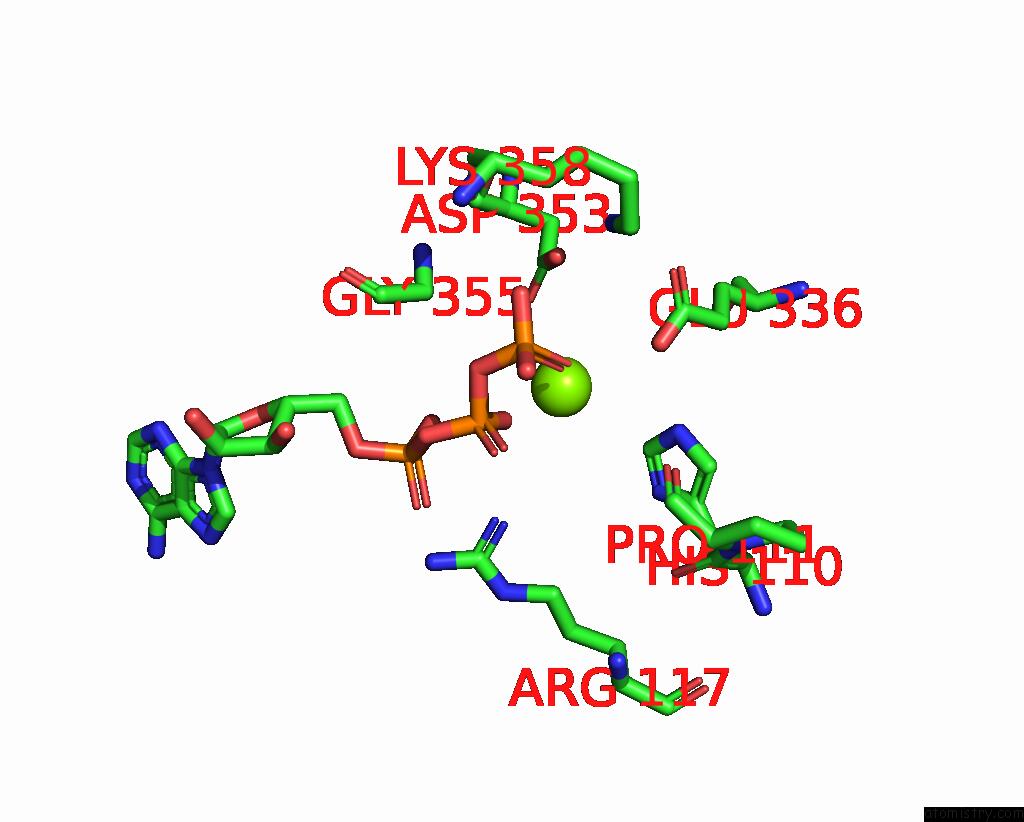
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 1 of Structure of ARP4-IES4-N-Actin-ARP8-INO80HSA Subcomplex (A-Module) of Chaetomium Thermophilum INO80 on Curved Dna within 5.0Å range:
|
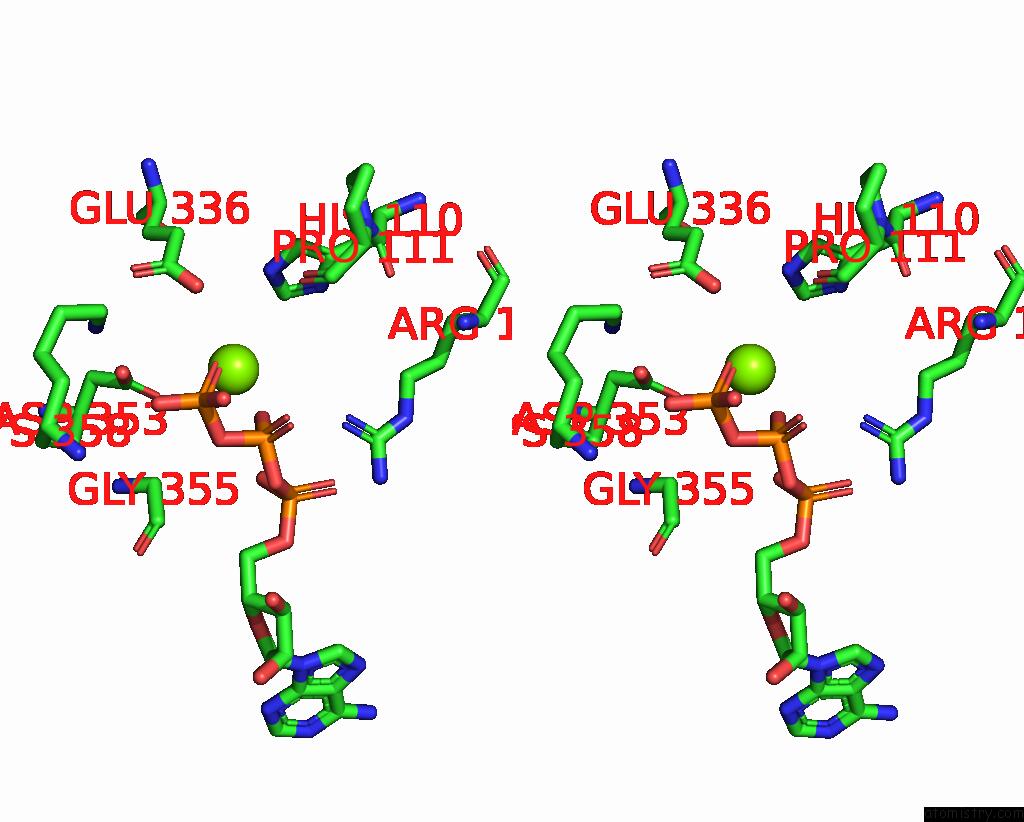
Magnesium binding site 2 out of 3 in 8a5p
Go back to
Magnesium binding site 2 out
of 3 in the Structure of ARP4-IES4-N-Actin-ARP8-INO80HSA Subcomplex (A-Module) of Chaetomium Thermophilum INO80 on Curved Dna

Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 2 of Structure of ARP4-IES4-N-Actin-ARP8-INO80HSA Subcomplex (A-Module) of Chaetomium Thermophilum INO80 on Curved Dna within 5.0Å range:
|
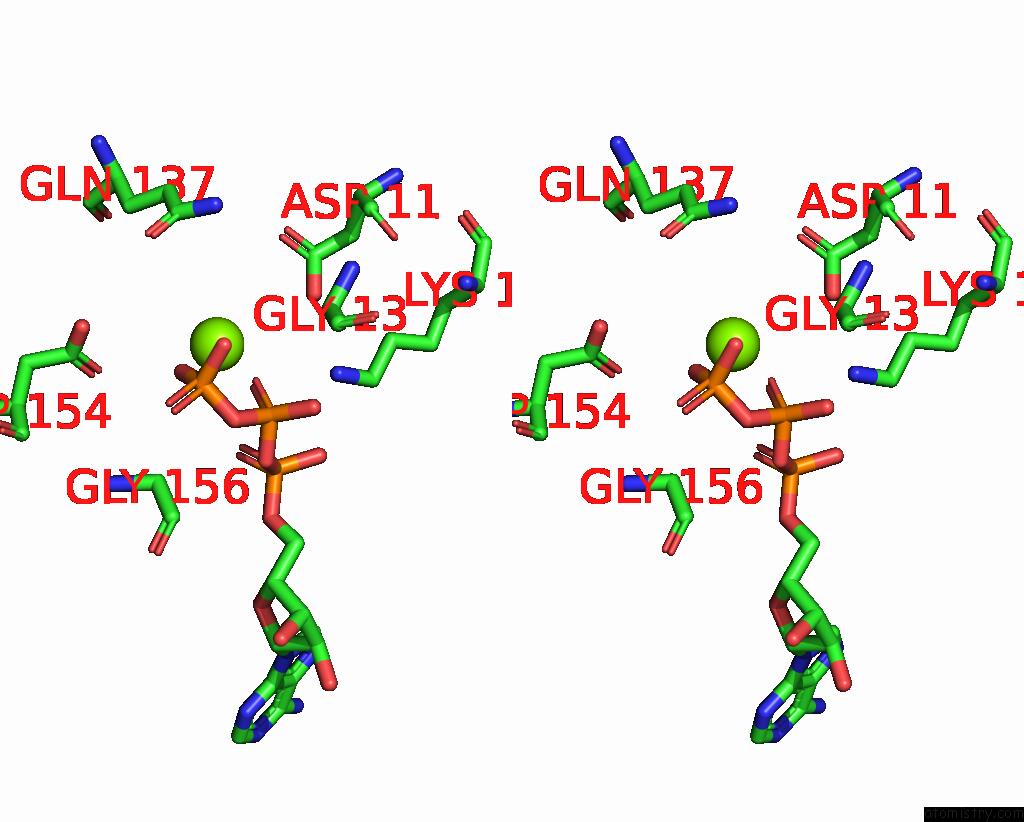
Magnesium binding site 3 out of 3 in 8a5p
Go back to
Magnesium binding site 3 out
of 3 in the Structure of ARP4-IES4-N-Actin-ARP8-INO80HSA Subcomplex (A-Module) of Chaetomium Thermophilum INO80 on Curved Dna
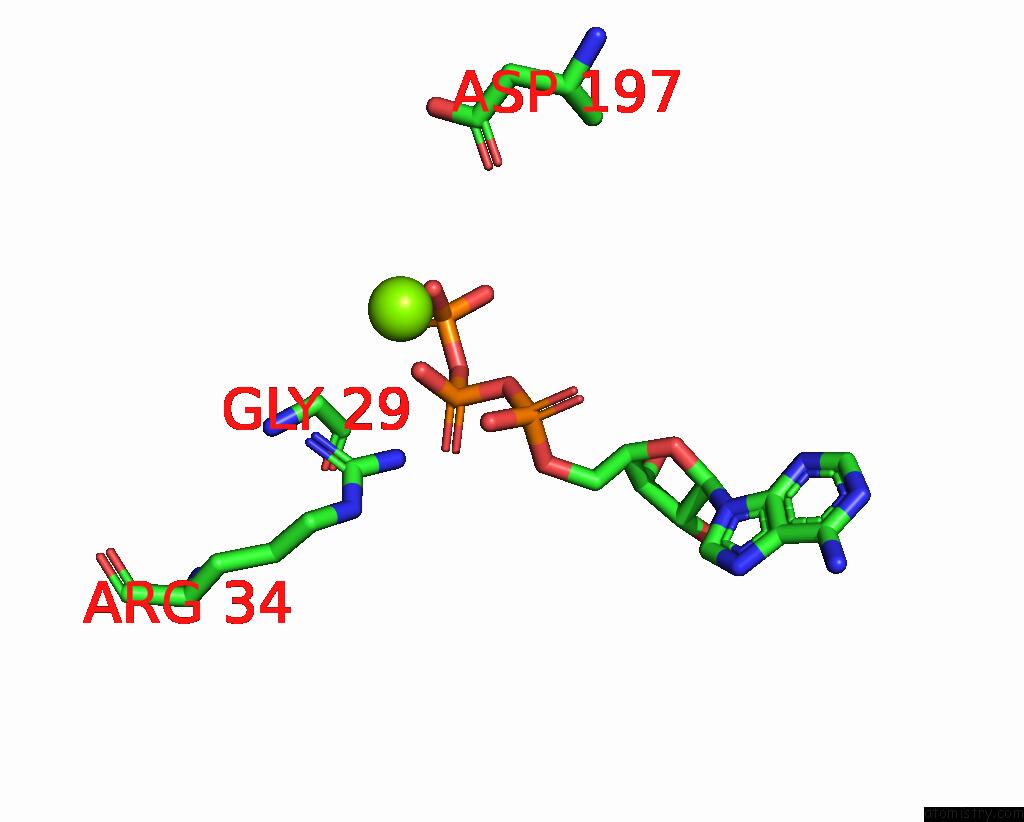
Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 3 of Structure of ARP4-IES4-N-Actin-ARP8-INO80HSA Subcomplex (A-Module) of Chaetomium Thermophilum INO80 on Curved Dna within 5.0Å range:
|
Reference:
F.Kunert,
F.J.Metzner,
J.Jung,
M.Hopfler,
S.Woike,
K.Schall,
D.Kostrewa,
M.Moldt,
J.X.Chen,
S.Bantele,
B.Pfander,
S.Eustermann,
K.P.Hopfner.
Structural Mechanism of Extranucleosomal Dna Readout By the INO80 Complex. Sci Adv V. 8 D3189 2022.
ISSN: ESSN 2375-2548
PubMed: 36490333
DOI: 10.1126/SCIADV.ADD3189
Page generated: Thu Oct 3 17:16:52 2024
ISSN: ESSN 2375-2548
PubMed: 36490333
DOI: 10.1126/SCIADV.ADD3189
Last articles
Al in 8SGCAl in 8SG9
Al in 8SFF
Al in 8SG8
Al in 8R1A
Al in 8Q75
Al in 8OIE
Al in 8OX6
Al in 8OX5
Al in 8OP8