Magnesium »
PDB 8acg-8aom »
8alr »
Magnesium in PDB 8alr: Small Molecular Stabilizer For Eralpha and 14-3-3 (1080272)
Protein crystallography data
The structure of Small Molecular Stabilizer For Eralpha and 14-3-3 (1080272), PDB code: 8alr
was solved by
E.J.Visser,
E.M.F.Vandenboorn,
C.Ottmann,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 45.42 / 1.40 |
| Space group | C 2 2 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 81.735, 112.169, 62.445, 90, 90, 90 |
| R / Rfree (%) | 14.8 / 17.1 |
Magnesium Binding Sites:
The binding sites of Magnesium atom in the Small Molecular Stabilizer For Eralpha and 14-3-3 (1080272)
(pdb code 8alr). This binding sites where shown within
5.0 Angstroms radius around Magnesium atom.
In total 2 binding sites of Magnesium where determined in the Small Molecular Stabilizer For Eralpha and 14-3-3 (1080272), PDB code: 8alr:
Jump to Magnesium binding site number: 1; 2;
In total 2 binding sites of Magnesium where determined in the Small Molecular Stabilizer For Eralpha and 14-3-3 (1080272), PDB code: 8alr:
Jump to Magnesium binding site number: 1; 2;
Magnesium binding site 1 out of 2 in 8alr
Go back to
Magnesium binding site 1 out
of 2 in the Small Molecular Stabilizer For Eralpha and 14-3-3 (1080272)
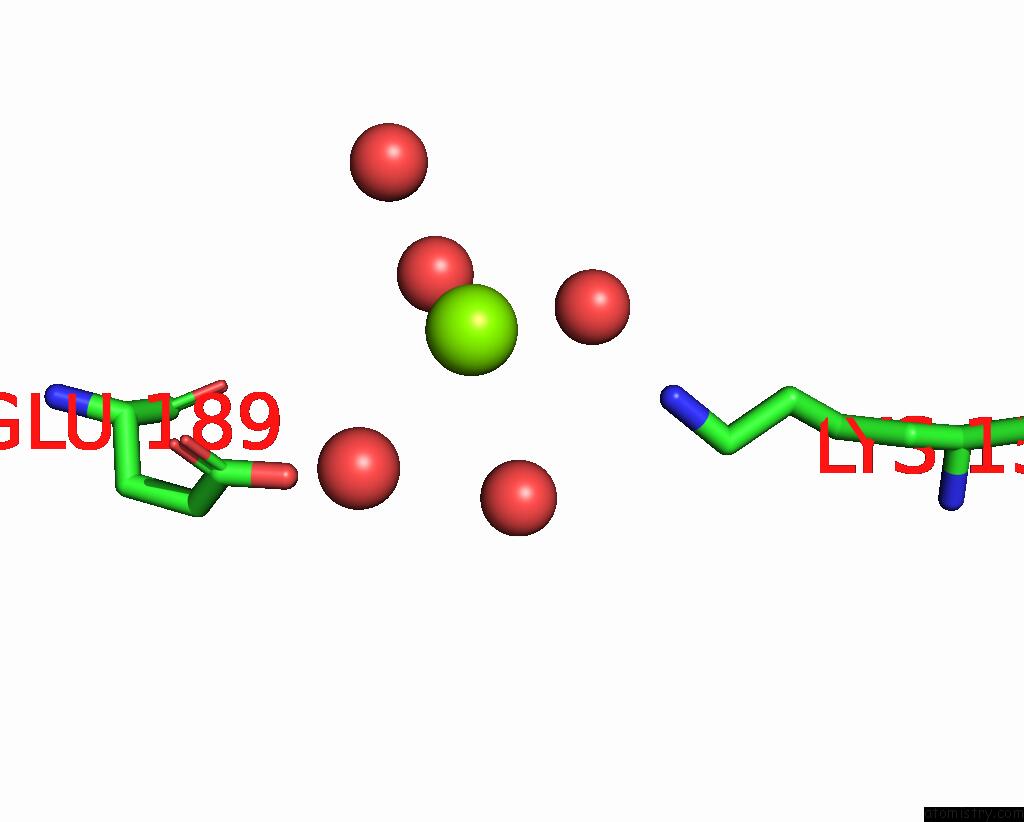
Mono view
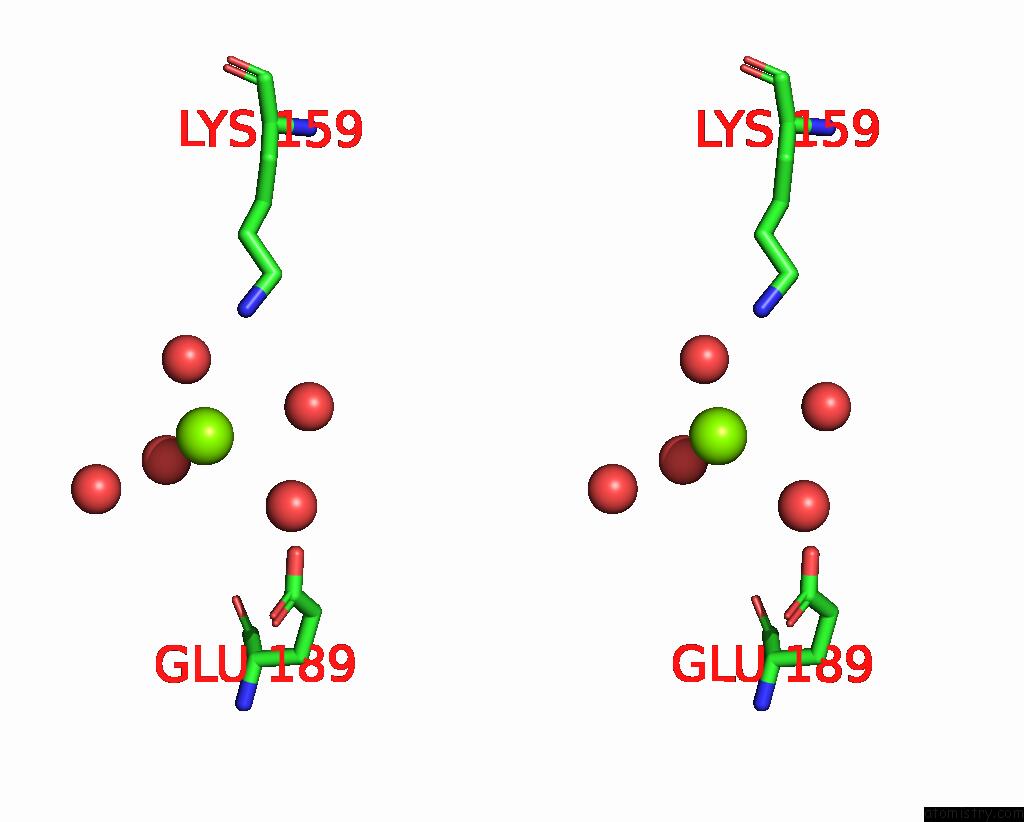
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 1 of Small Molecular Stabilizer For Eralpha and 14-3-3 (1080272) within 5.0Å range:
|
Magnesium binding site 2 out of 2 in 8alr
Go back to
Magnesium binding site 2 out
of 2 in the Small Molecular Stabilizer For Eralpha and 14-3-3 (1080272)
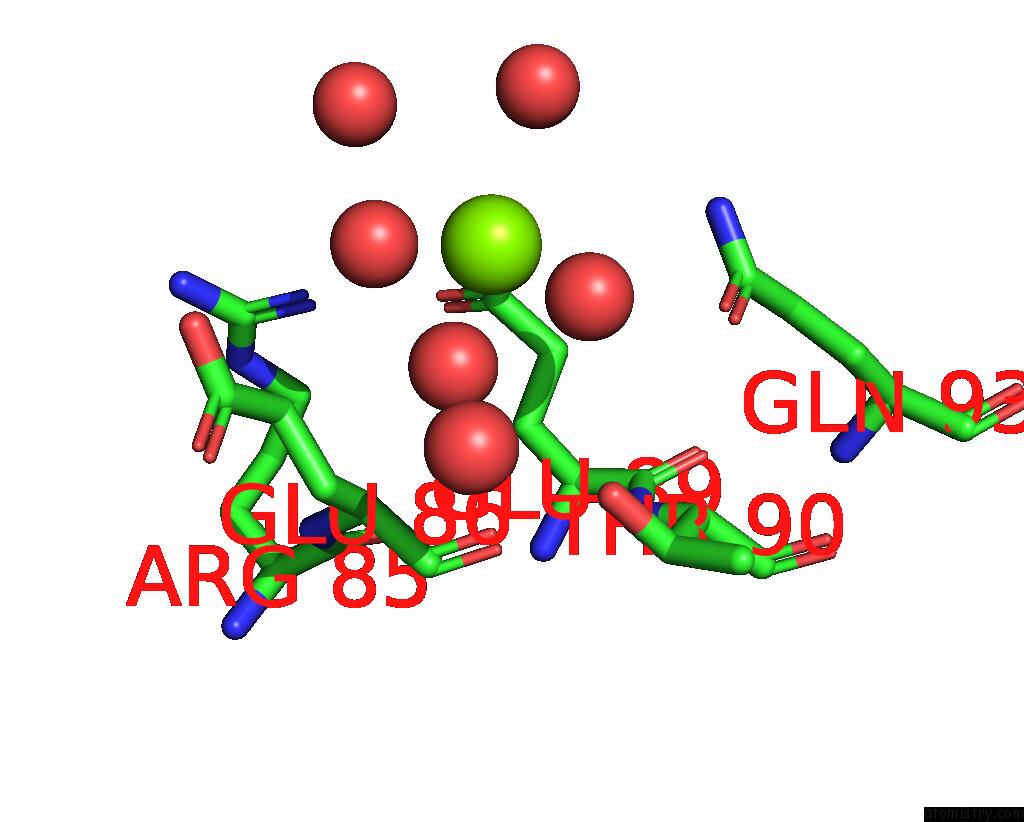
Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 2 of Small Molecular Stabilizer For Eralpha and 14-3-3 (1080272) within 5.0Å range:
|
Reference:
M.Konstantinidou,
E.J.Visser,
E.Vandenboorn,
S.Chen,
P.Jaishankar,
M.Overmans,
S.Dutta,
R.J.Neitz,
A.R.Renslo,
C.Ottmann,
L.Brunsveld,
M.R.Arkin.
Structure-Based Optimization of Covalent, Small-Molecule Stabilizers of the 14-3-3 Sigma /Er Alpha Protein-Protein Interaction From Nonselective Fragments. J.Am.Chem.Soc. 2023.
ISSN: ESSN 1520-5126
PubMed: 37676236
DOI: 10.1021/JACS.3C05161
Page generated: Thu Oct 3 18:08:56 2024
ISSN: ESSN 1520-5126
PubMed: 37676236
DOI: 10.1021/JACS.3C05161
Last articles
F in 4FXYF in 4FXQ
F in 4FZF
F in 4FP1
F in 4FVX
F in 4FV1
F in 4FV9
F in 4FV3
F in 4FS2
F in 4FV0