Magnesium »
PDB 8dh6-8dr5 »
8dqv »
Magnesium in PDB 8dqv: The 1.52 Angstrom Cryoem Structure of the [Nife]-Hydrogenase Huc From Mycobacterium Smegmatis - Catalytic Dimer (HUC2S2L)
Enzymatic activity of The 1.52 Angstrom Cryoem Structure of the [Nife]-Hydrogenase Huc From Mycobacterium Smegmatis - Catalytic Dimer (HUC2S2L)
All present enzymatic activity of The 1.52 Angstrom Cryoem Structure of the [Nife]-Hydrogenase Huc From Mycobacterium Smegmatis - Catalytic Dimer (HUC2S2L):
1.12.99.6;
1.12.99.6;
Other elements in 8dqv:
The structure of The 1.52 Angstrom Cryoem Structure of the [Nife]-Hydrogenase Huc From Mycobacterium Smegmatis - Catalytic Dimer (HUC2S2L) also contains other interesting chemical elements:
| Iron | (Fe) | 20 atoms |
| Nickel | (Ni) | 2 atoms |
Magnesium Binding Sites:
The binding sites of Magnesium atom in the The 1.52 Angstrom Cryoem Structure of the [Nife]-Hydrogenase Huc From Mycobacterium Smegmatis - Catalytic Dimer (HUC2S2L)
(pdb code 8dqv). This binding sites where shown within
5.0 Angstroms radius around Magnesium atom.
In total 2 binding sites of Magnesium where determined in the The 1.52 Angstrom Cryoem Structure of the [Nife]-Hydrogenase Huc From Mycobacterium Smegmatis - Catalytic Dimer (HUC2S2L), PDB code: 8dqv:
Jump to Magnesium binding site number: 1; 2;
In total 2 binding sites of Magnesium where determined in the The 1.52 Angstrom Cryoem Structure of the [Nife]-Hydrogenase Huc From Mycobacterium Smegmatis - Catalytic Dimer (HUC2S2L), PDB code: 8dqv:
Jump to Magnesium binding site number: 1; 2;
Magnesium binding site 1 out of 2 in 8dqv
Go back to
Magnesium binding site 1 out
of 2 in the The 1.52 Angstrom Cryoem Structure of the [Nife]-Hydrogenase Huc From Mycobacterium Smegmatis - Catalytic Dimer (HUC2S2L)
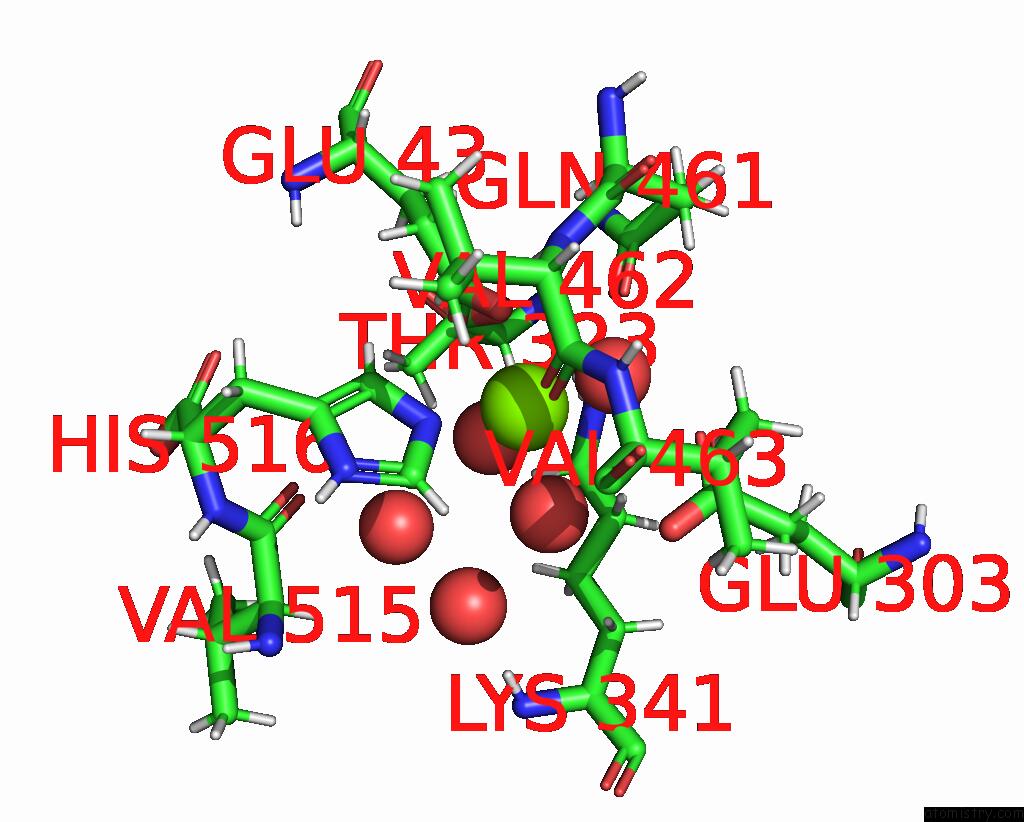
Mono view
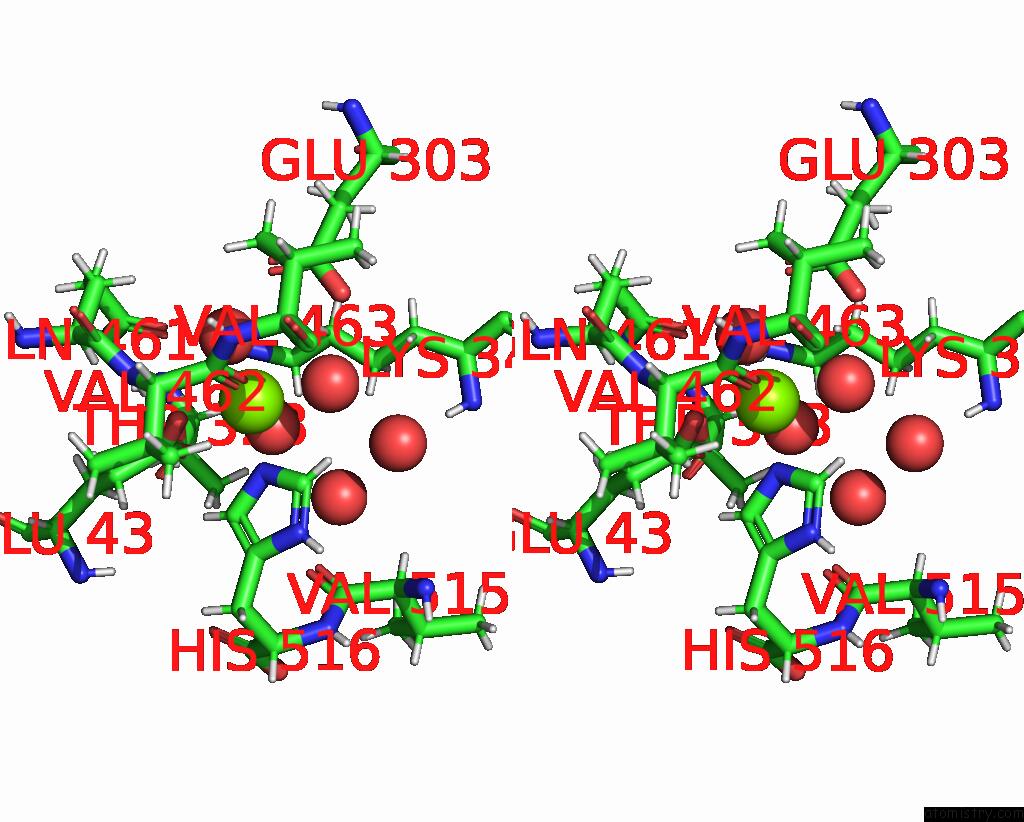
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 1 of The 1.52 Angstrom Cryoem Structure of the [Nife]-Hydrogenase Huc From Mycobacterium Smegmatis - Catalytic Dimer (HUC2S2L) within 5.0Å range:
|
Magnesium binding site 2 out of 2 in 8dqv
Go back to
Magnesium binding site 2 out
of 2 in the The 1.52 Angstrom Cryoem Structure of the [Nife]-Hydrogenase Huc From Mycobacterium Smegmatis - Catalytic Dimer (HUC2S2L)
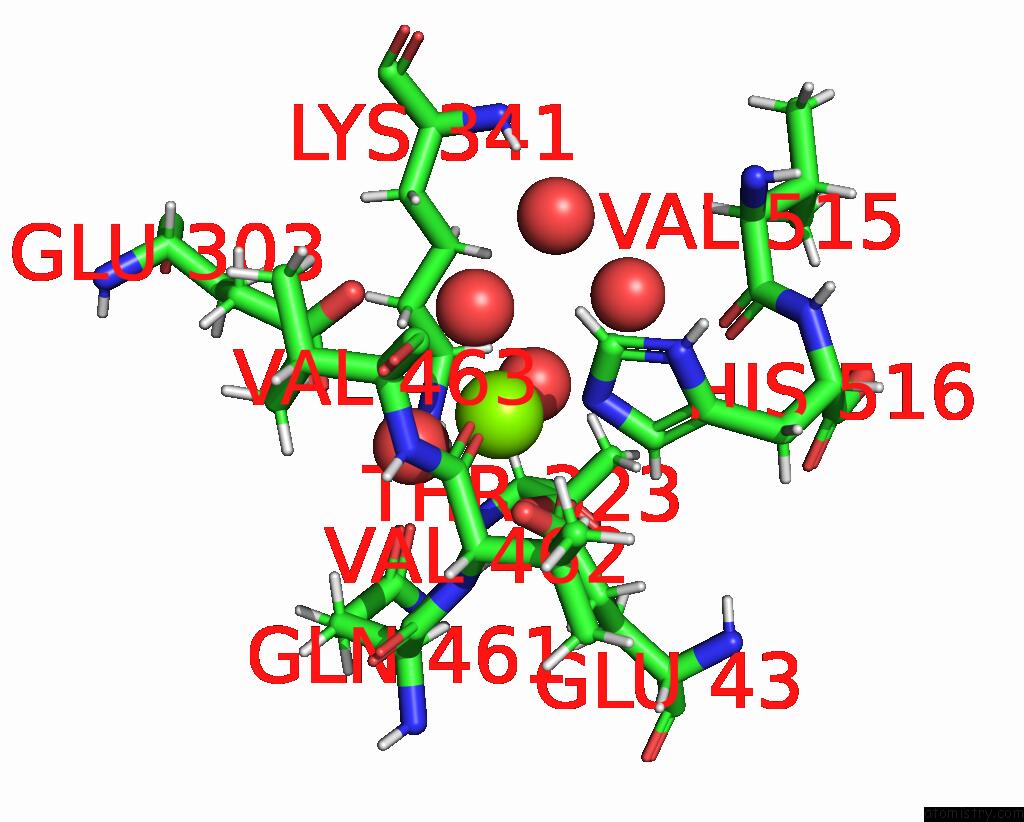
Mono view
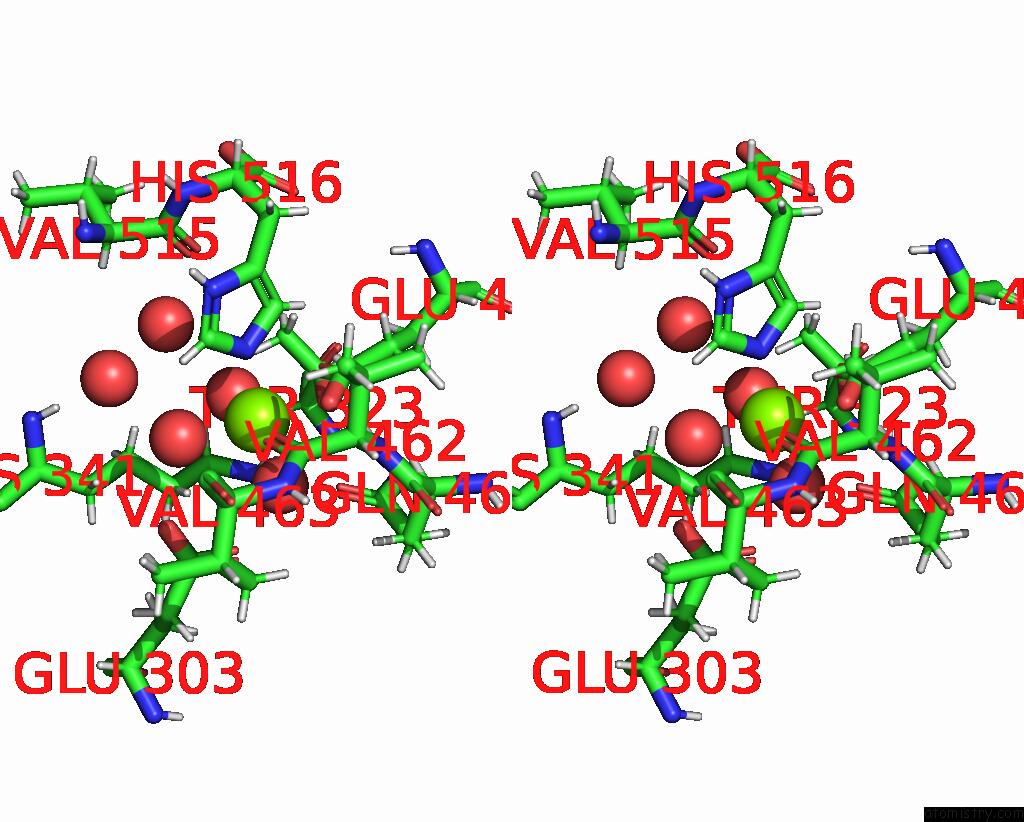
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 2 of The 1.52 Angstrom Cryoem Structure of the [Nife]-Hydrogenase Huc From Mycobacterium Smegmatis - Catalytic Dimer (HUC2S2L) within 5.0Å range:
|
Reference:
R.Grinter,
A.Kropp,
H.Venugopal,
M.Senger,
J.Badley,
P.Cabotaje,
S.T.Stripp,
C.K.Barlow,
M.Belousoff,
G.M.Cook,
R.B.Schittenhelm,
S.Khalid,
G.Berggren,
G.Greening.
Energy Extraction From Air: Structural Basis of Atmospheric Hydrogen Oxidation To Be Published.
Page generated: Fri Oct 4 00:48:37 2024
Last articles
Ca in 5T3HCa in 5T2N
Ca in 5T2O
Ca in 5T2H
Ca in 5SZQ
Ca in 5SZO
Ca in 5SZP
Ca in 5SZN
Ca in 5SZM
Ca in 5SZL