Magnesium »
PDB 8fiy-8ful »
8fmj »
Magnesium in PDB 8fmj: Crystal Structure of Human Kras in Space Group R32
Enzymatic activity of Crystal Structure of Human Kras in Space Group R32
All present enzymatic activity of Crystal Structure of Human Kras in Space Group R32:
3.6.5.2;
3.6.5.2;
Protein crystallography data
The structure of Crystal Structure of Human Kras in Space Group R32, PDB code: 8fmj
was solved by
R.Brenner,
A.Landgraf,
G.Gonzalez-Gutierrez,
K.Bum-Erdene,
S.O.Meroueh,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 47.45 / 1.33 |
| Space group | H 3 2 |
| Cell size a, b, c (Å), α, β, γ (°) | 92.516, 92.516, 117.775, 90, 90, 120 |
| R / Rfree (%) | 14.2 / 16.7 |
Magnesium Binding Sites:
The binding sites of Magnesium atom in the Crystal Structure of Human Kras in Space Group R32
(pdb code 8fmj). This binding sites where shown within
5.0 Angstroms radius around Magnesium atom.
In total only one binding site of Magnesium was determined in the Crystal Structure of Human Kras in Space Group R32, PDB code: 8fmj:
In total only one binding site of Magnesium was determined in the Crystal Structure of Human Kras in Space Group R32, PDB code: 8fmj:
Magnesium binding site 1 out of 1 in 8fmj
Go back to
Magnesium binding site 1 out
of 1 in the Crystal Structure of Human Kras in Space Group R32
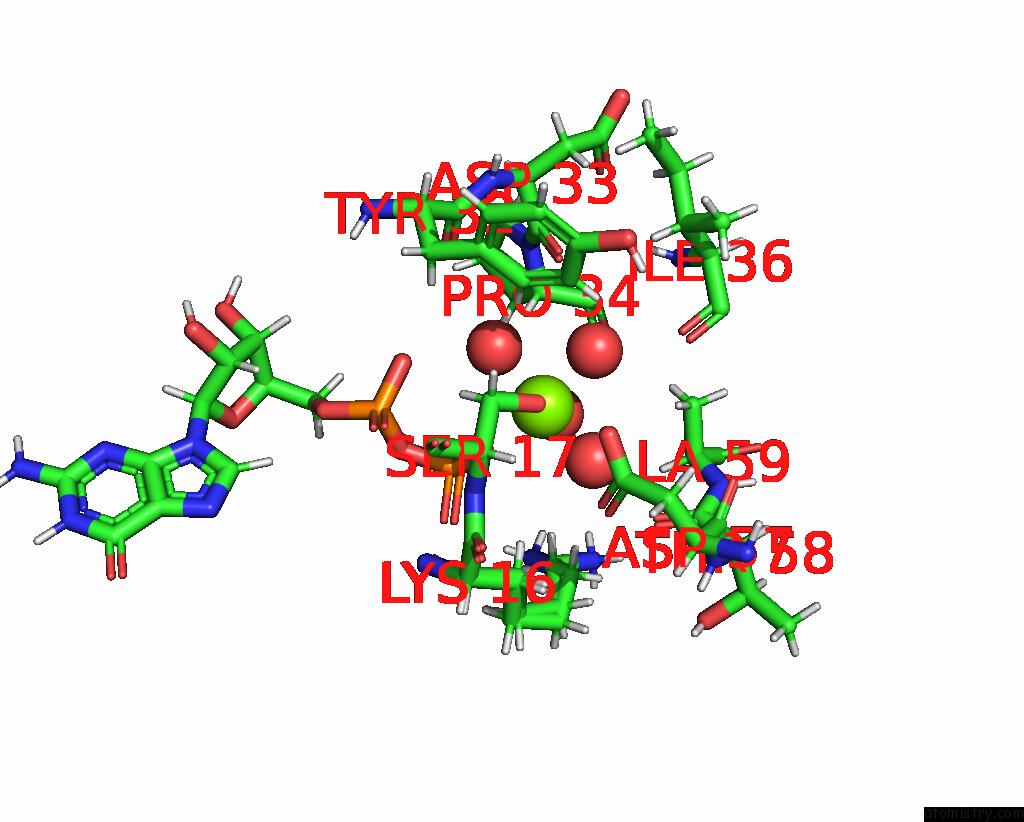
Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 1 of Crystal Structure of Human Kras in Space Group R32 within 5.0Å range:
|
Reference:
R.J.Brenner,
A.D.Landgraf,
K.Bum-Erdene,
G.Gonzalez-Gutierrez,
S.O.Meroueh.
Crystal Packing Reveals A Potential Autoinhibited Kras Dimer Interface and A Strategy For Small-Molecule Inhibition of Ras Signaling. Biochemistry 2023.
ISSN: ISSN 0006-2960
PubMed: 37938120
DOI: 10.1021/ACS.BIOCHEM.3C00378
Page generated: Fri Aug 15 04:52:59 2025
ISSN: ISSN 0006-2960
PubMed: 37938120
DOI: 10.1021/ACS.BIOCHEM.3C00378
Last articles
Mg in 8HIMMg in 8HHL
Mg in 8HIL
Mg in 8HHM
Mg in 8HG6
Mg in 8HHA
Mg in 8HH9
Mg in 8HH8
Mg in 8HH6
Mg in 8HH5