Magnesium »
PDB 8q2z-8qbk »
8q7w »
Magnesium in PDB 8q7w: Structure of the Recycling U5 Snrnp Bound to Chaperone CD2BP2 (State 3)
Enzymatic activity of Structure of the Recycling U5 Snrnp Bound to Chaperone CD2BP2 (State 3)
All present enzymatic activity of Structure of the Recycling U5 Snrnp Bound to Chaperone CD2BP2 (State 3):
3.6.4.13;
3.6.4.13;
Magnesium Binding Sites:
The binding sites of Magnesium atom in the Structure of the Recycling U5 Snrnp Bound to Chaperone CD2BP2 (State 3)
(pdb code 8q7w). This binding sites where shown within
5.0 Angstroms radius around Magnesium atom.
In total only one binding site of Magnesium was determined in the Structure of the Recycling U5 Snrnp Bound to Chaperone CD2BP2 (State 3), PDB code: 8q7w:
In total only one binding site of Magnesium was determined in the Structure of the Recycling U5 Snrnp Bound to Chaperone CD2BP2 (State 3), PDB code: 8q7w:
Magnesium binding site 1 out of 1 in 8q7w
Go back to
Magnesium binding site 1 out
of 1 in the Structure of the Recycling U5 Snrnp Bound to Chaperone CD2BP2 (State 3)
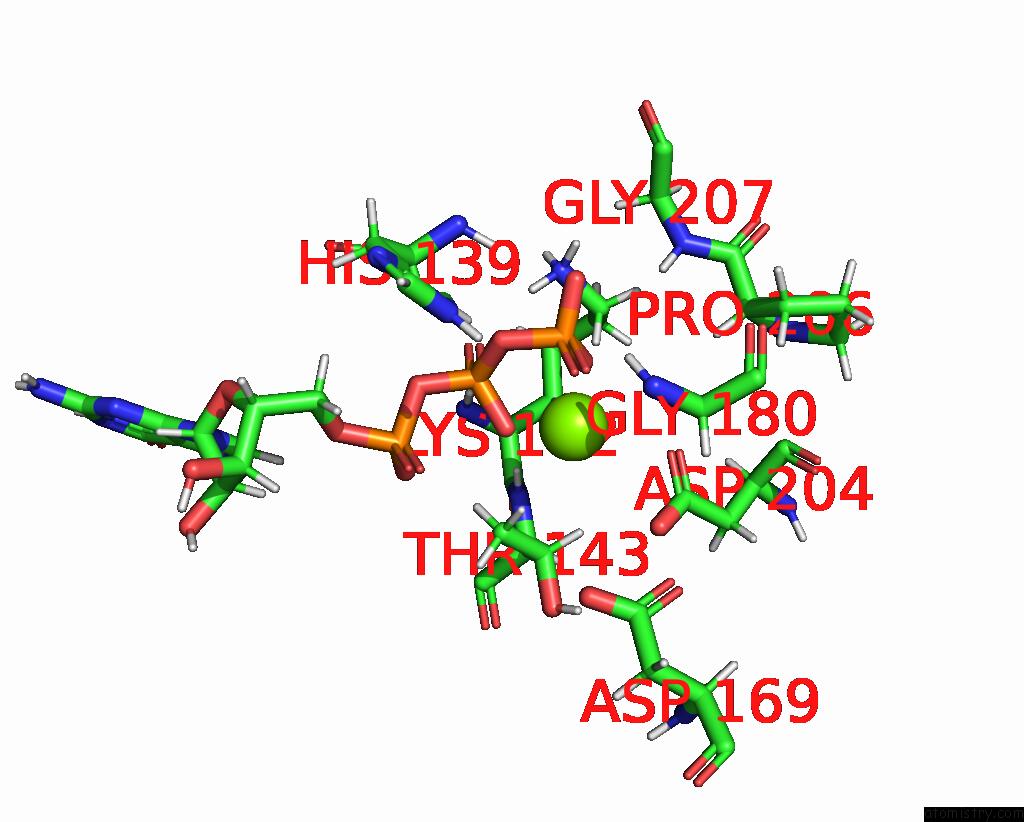
Mono view
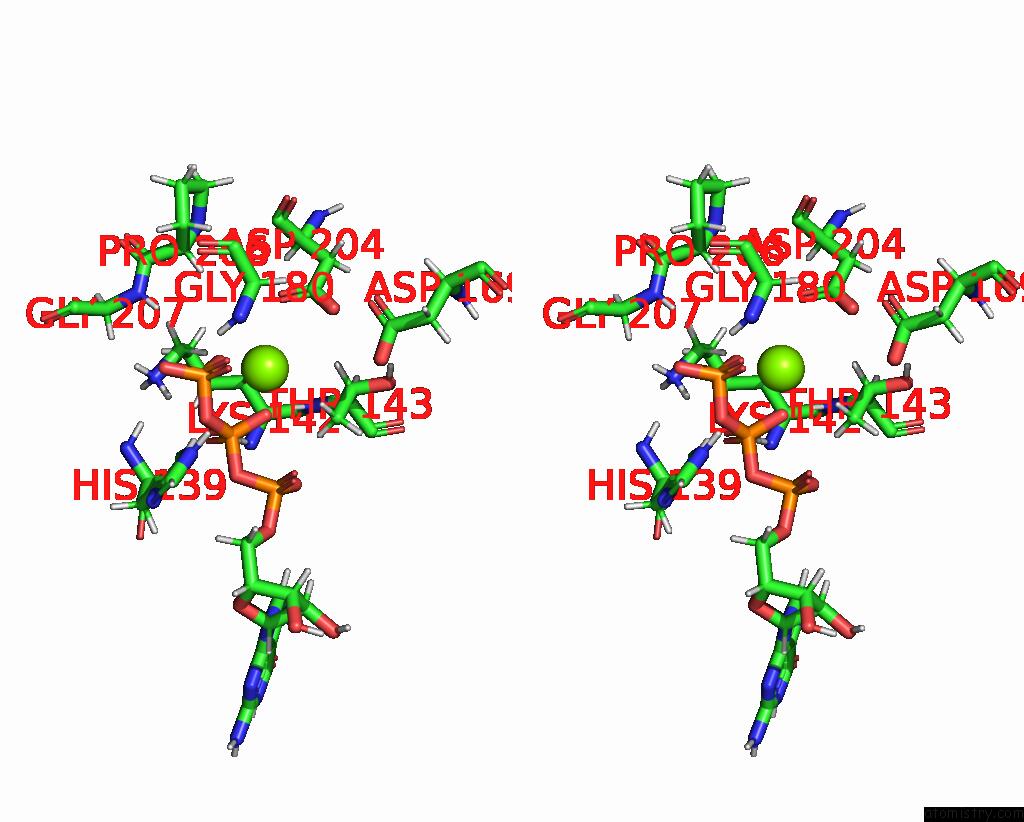
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 1 of Structure of the Recycling U5 Snrnp Bound to Chaperone CD2BP2 (State 3) within 5.0Å range:
|
Reference:
D.Riabov Bassat,
S.Visanpattanasin,
M.K.Vorlander,
L.Fin,
A.W.Phillips,
C.Plaschka.
Structural Basis of Human U5 Snrnp Late Biogenesis and Recycling. Nat.Struct.Mol.Biol. 2024.
ISSN: ESSN 1545-9985
PubMed: 38467876
DOI: 10.1038/S41594-024-01243-4
Page generated: Fri Oct 4 16:39:48 2024
ISSN: ESSN 1545-9985
PubMed: 38467876
DOI: 10.1038/S41594-024-01243-4
Last articles
F in 4HUAF in 4HU9
F in 4HQJ
F in 4HT3
F in 4HLQ
F in 4HT0
F in 4HNA
F in 4HPX
F in 4HQH
F in 4HNS