Magnesium »
PDB 8qla-8qs7 »
8qmw »
Magnesium in PDB 8qmw: Non-Obligately L8S8-Complex Forming Rubisco Derived From Ancestral Sequence Reconstruction and Rational Engineering in L8S8 Complex with Substitutions R269W, E271R, L273N
Enzymatic activity of Non-Obligately L8S8-Complex Forming Rubisco Derived From Ancestral Sequence Reconstruction and Rational Engineering in L8S8 Complex with Substitutions R269W, E271R, L273N
All present enzymatic activity of Non-Obligately L8S8-Complex Forming Rubisco Derived From Ancestral Sequence Reconstruction and Rational Engineering in L8S8 Complex with Substitutions R269W, E271R, L273N:
4.1.1.39;
4.1.1.39;
Protein crystallography data
The structure of Non-Obligately L8S8-Complex Forming Rubisco Derived From Ancestral Sequence Reconstruction and Rational Engineering in L8S8 Complex with Substitutions R269W, E271R, L273N, PDB code: 8qmw
was solved by
J.Zarzycki,
L.Schulz,
T.J.Erb,
G.K.A.Hochberg,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 24.88 / 1.75 |
| Space group | C 1 2 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 206.01, 106.44, 108.49, 90, 113.09, 90 |
| R / Rfree (%) | 14.5 / 17.5 |
Magnesium Binding Sites:
The binding sites of Magnesium atom in the Non-Obligately L8S8-Complex Forming Rubisco Derived From Ancestral Sequence Reconstruction and Rational Engineering in L8S8 Complex with Substitutions R269W, E271R, L273N
(pdb code 8qmw). This binding sites where shown within
5.0 Angstroms radius around Magnesium atom.
In total 4 binding sites of Magnesium where determined in the Non-Obligately L8S8-Complex Forming Rubisco Derived From Ancestral Sequence Reconstruction and Rational Engineering in L8S8 Complex with Substitutions R269W, E271R, L273N, PDB code: 8qmw:
Jump to Magnesium binding site number: 1; 2; 3; 4;
In total 4 binding sites of Magnesium where determined in the Non-Obligately L8S8-Complex Forming Rubisco Derived From Ancestral Sequence Reconstruction and Rational Engineering in L8S8 Complex with Substitutions R269W, E271R, L273N, PDB code: 8qmw:
Jump to Magnesium binding site number: 1; 2; 3; 4;
Magnesium binding site 1 out of 4 in 8qmw
Go back to
Magnesium binding site 1 out
of 4 in the Non-Obligately L8S8-Complex Forming Rubisco Derived From Ancestral Sequence Reconstruction and Rational Engineering in L8S8 Complex with Substitutions R269W, E271R, L273N
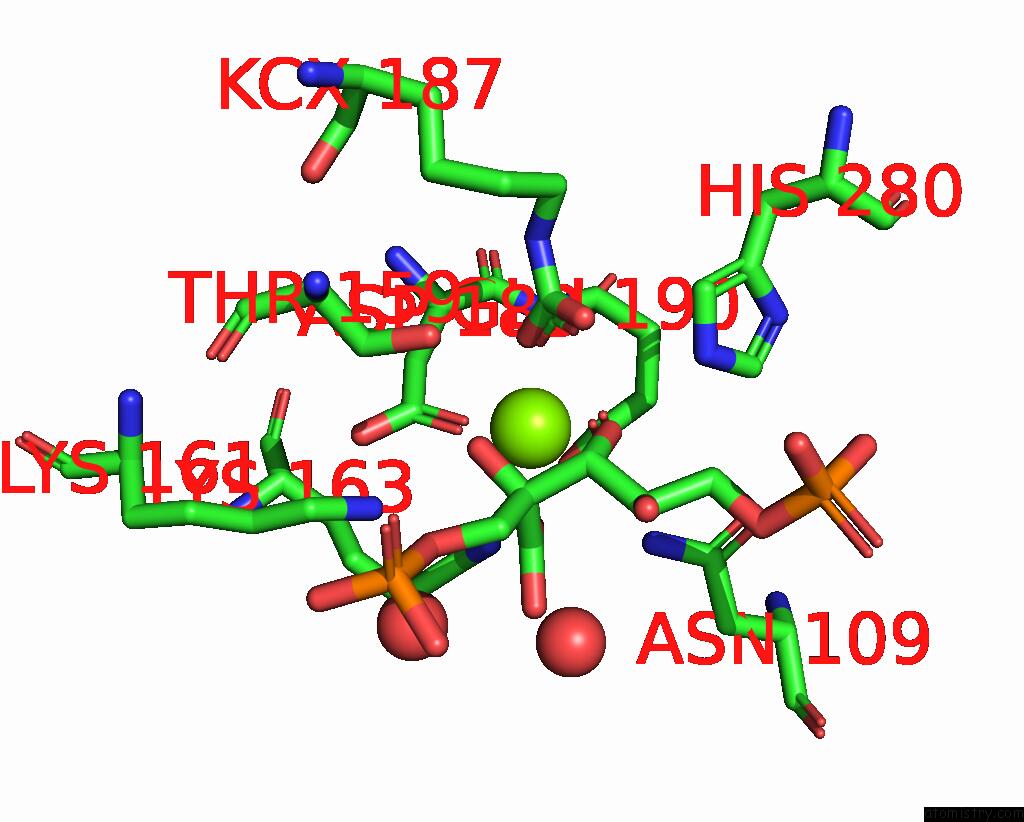
Mono view
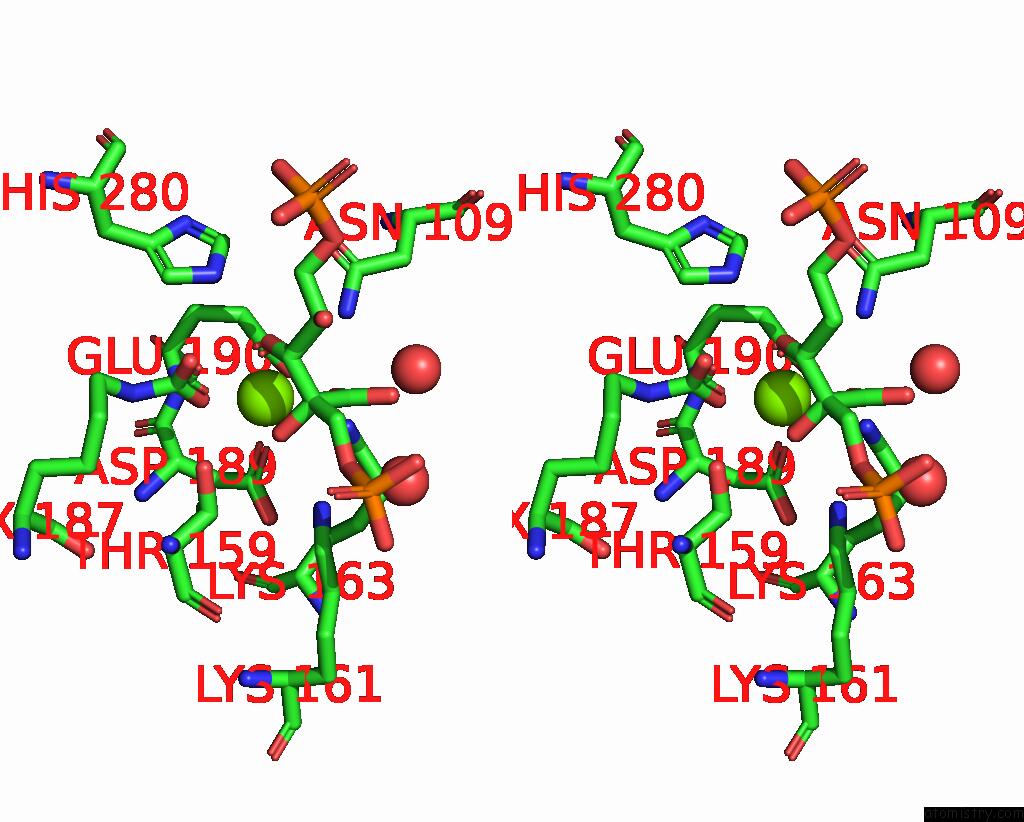
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 1 of Non-Obligately L8S8-Complex Forming Rubisco Derived From Ancestral Sequence Reconstruction and Rational Engineering in L8S8 Complex with Substitutions R269W, E271R, L273N within 5.0Å range:
|
Magnesium binding site 2 out of 4 in 8qmw
Go back to
Magnesium binding site 2 out
of 4 in the Non-Obligately L8S8-Complex Forming Rubisco Derived From Ancestral Sequence Reconstruction and Rational Engineering in L8S8 Complex with Substitutions R269W, E271R, L273N
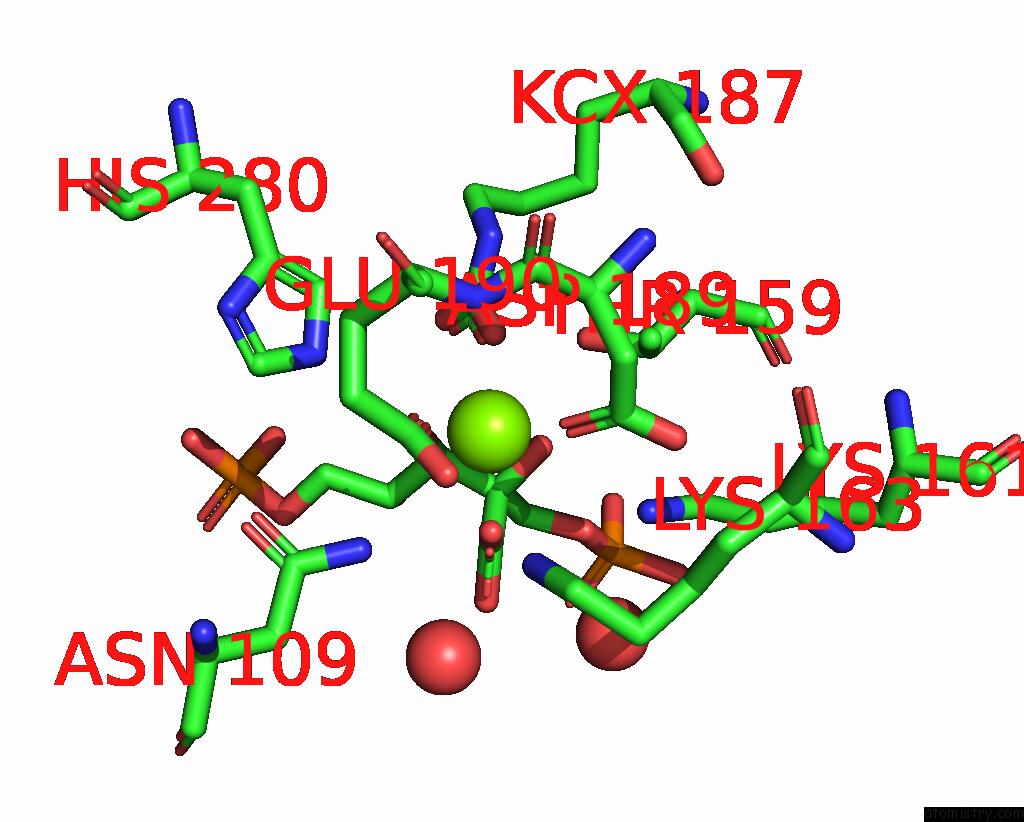
Mono view
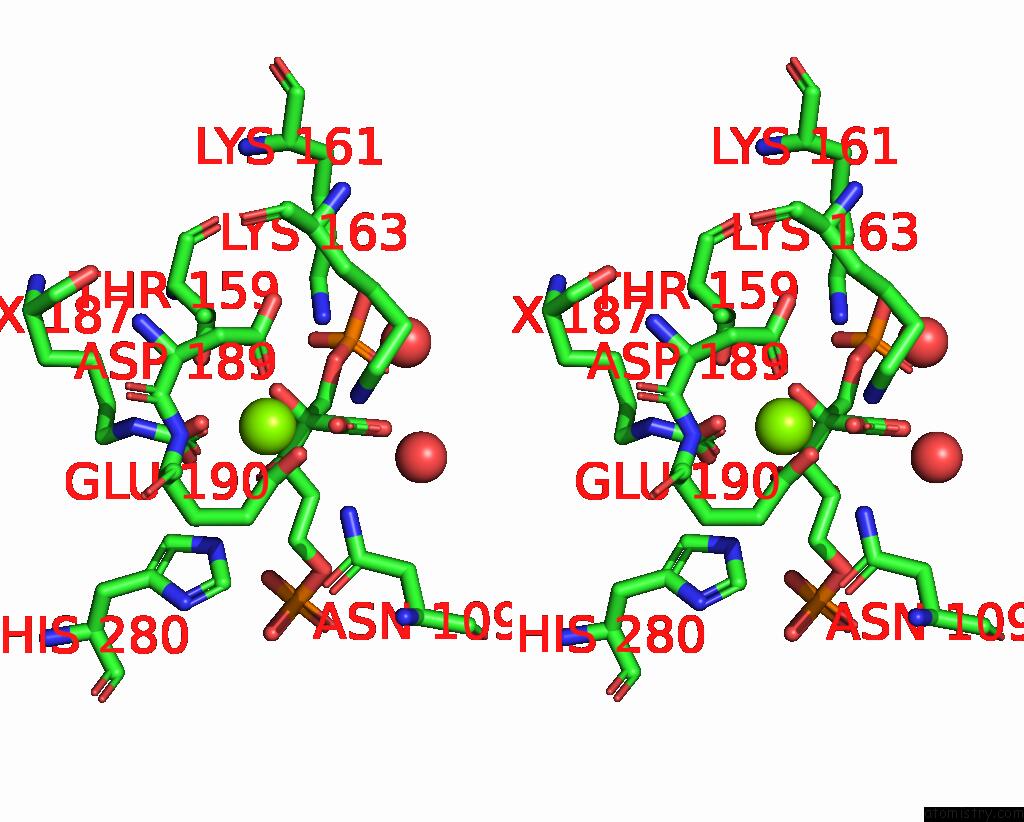
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 2 of Non-Obligately L8S8-Complex Forming Rubisco Derived From Ancestral Sequence Reconstruction and Rational Engineering in L8S8 Complex with Substitutions R269W, E271R, L273N within 5.0Å range:
|
Magnesium binding site 3 out of 4 in 8qmw
Go back to
Magnesium binding site 3 out
of 4 in the Non-Obligately L8S8-Complex Forming Rubisco Derived From Ancestral Sequence Reconstruction and Rational Engineering in L8S8 Complex with Substitutions R269W, E271R, L273N
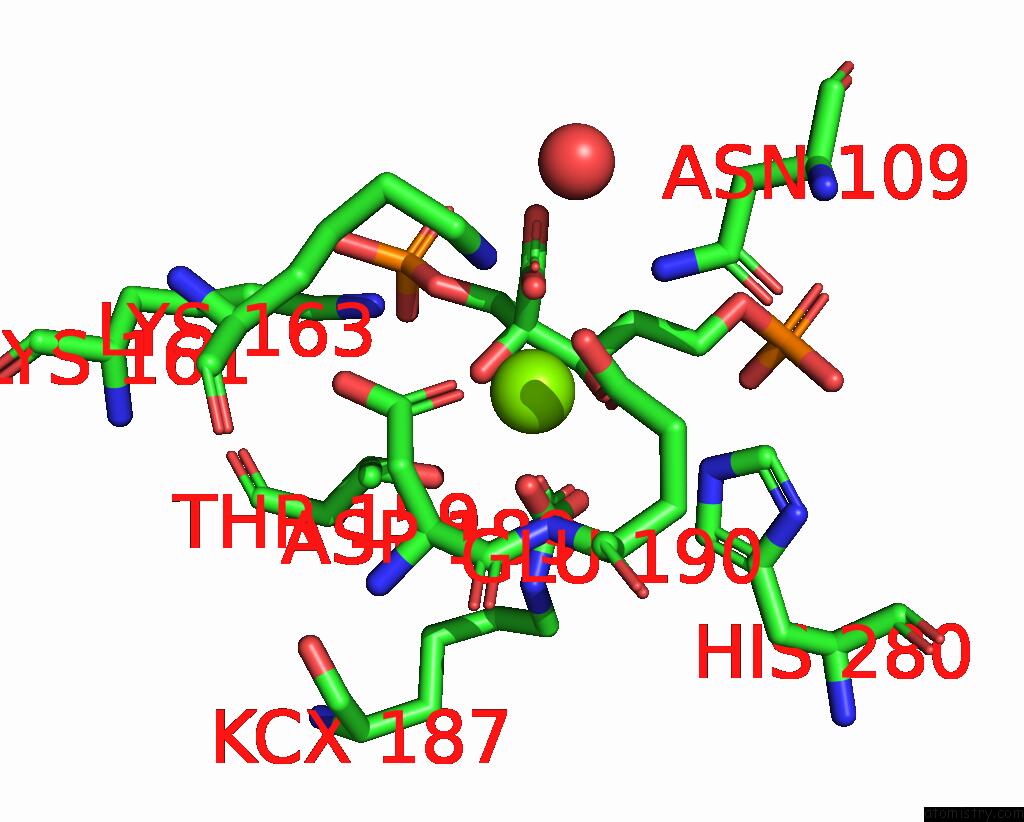
Mono view
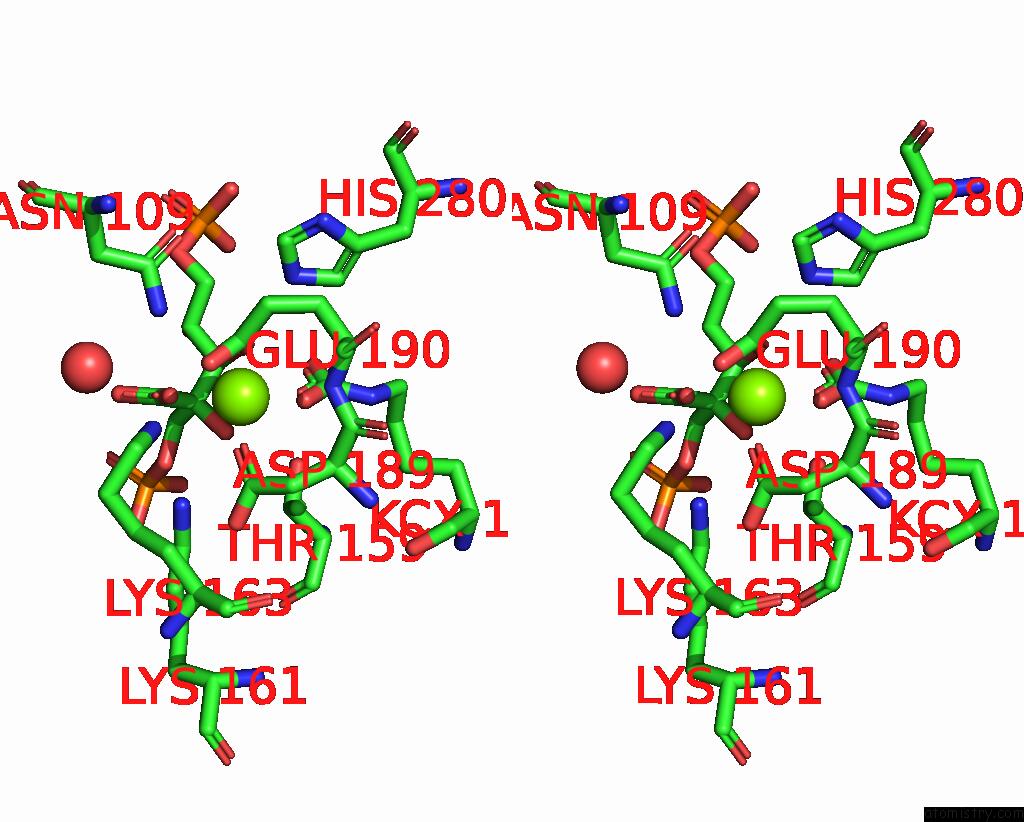
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 3 of Non-Obligately L8S8-Complex Forming Rubisco Derived From Ancestral Sequence Reconstruction and Rational Engineering in L8S8 Complex with Substitutions R269W, E271R, L273N within 5.0Å range:
|
Magnesium binding site 4 out of 4 in 8qmw
Go back to
Magnesium binding site 4 out
of 4 in the Non-Obligately L8S8-Complex Forming Rubisco Derived From Ancestral Sequence Reconstruction and Rational Engineering in L8S8 Complex with Substitutions R269W, E271R, L273N
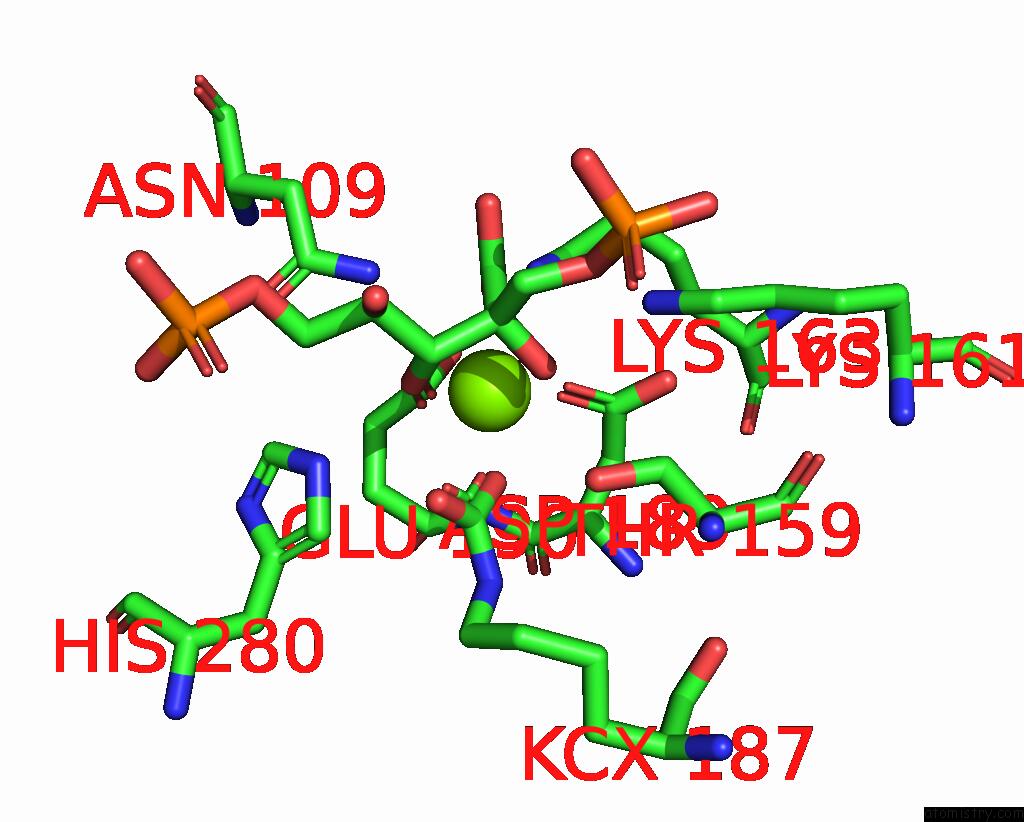
Mono view
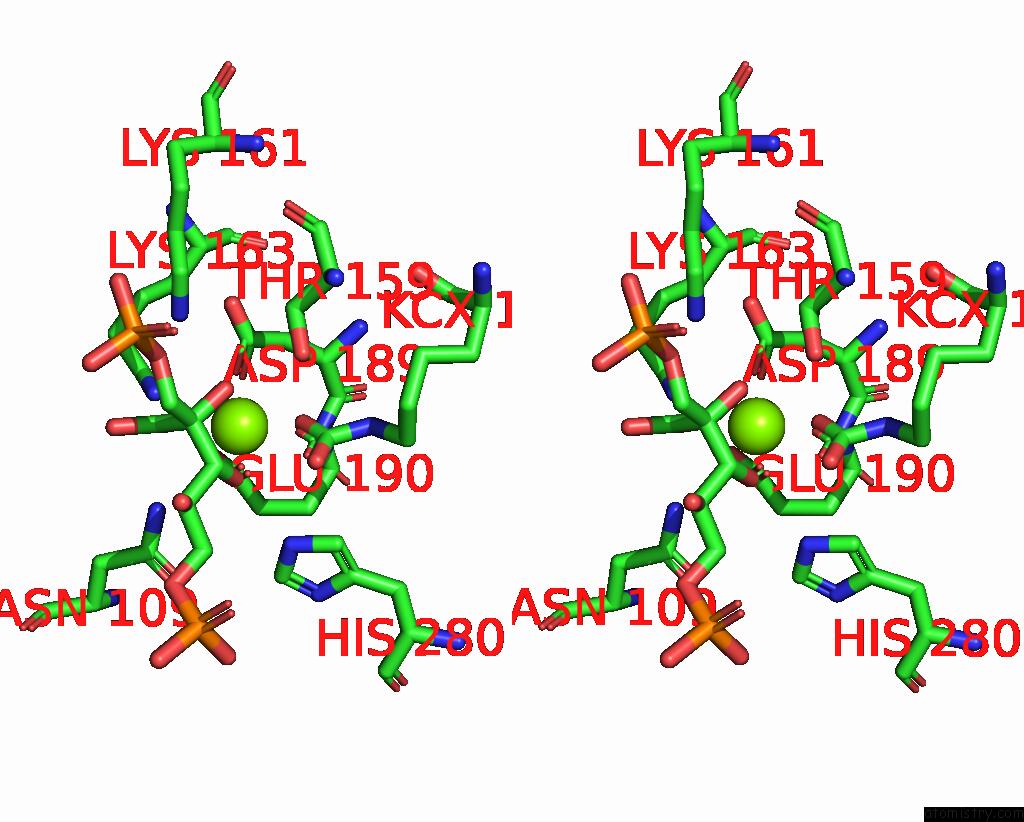
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 4 of Non-Obligately L8S8-Complex Forming Rubisco Derived From Ancestral Sequence Reconstruction and Rational Engineering in L8S8 Complex with Substitutions R269W, E271R, L273N within 5.0Å range:
|
Reference:
L.Schulz,
J.Zarzycki,
W.Steinchen,
T.J.Erb,
G.K.A.Hochberg.
Layered Entrenchment Maintains Essentiality in Protein-Protein Interactions Embo J. 2024.
ISSN: ESSN 1460-2075
Page generated: Fri Aug 15 13:12:45 2025
ISSN: ESSN 1460-2075
Last articles
Mg in 8S8KMg in 8S8G
Mg in 8S8D
Mg in 8S8E
Mg in 8S8F
Mg in 8S1P
Mg in 8RW1
Mg in 8S87
Mg in 8S8B
Mg in 8S8A