Magnesium »
PDB 8qla-8qs7 »
8qqi »
Magnesium in PDB 8qqi: E.Coli Dna Gyrase in Complex with 217 Bp Substrate Dna and Lei-800
Enzymatic activity of E.Coli Dna Gyrase in Complex with 217 Bp Substrate Dna and Lei-800
All present enzymatic activity of E.Coli Dna Gyrase in Complex with 217 Bp Substrate Dna and Lei-800:
5.6.2.2;
5.6.2.2;
Magnesium Binding Sites:
The binding sites of Magnesium atom in the E.Coli Dna Gyrase in Complex with 217 Bp Substrate Dna and Lei-800
(pdb code 8qqi). This binding sites where shown within
5.0 Angstroms radius around Magnesium atom.
In total 2 binding sites of Magnesium where determined in the E.Coli Dna Gyrase in Complex with 217 Bp Substrate Dna and Lei-800, PDB code: 8qqi:
Jump to Magnesium binding site number: 1; 2;
In total 2 binding sites of Magnesium where determined in the E.Coli Dna Gyrase in Complex with 217 Bp Substrate Dna and Lei-800, PDB code: 8qqi:
Jump to Magnesium binding site number: 1; 2;
Magnesium binding site 1 out of 2 in 8qqi
Go back to
Magnesium binding site 1 out
of 2 in the E.Coli Dna Gyrase in Complex with 217 Bp Substrate Dna and Lei-800
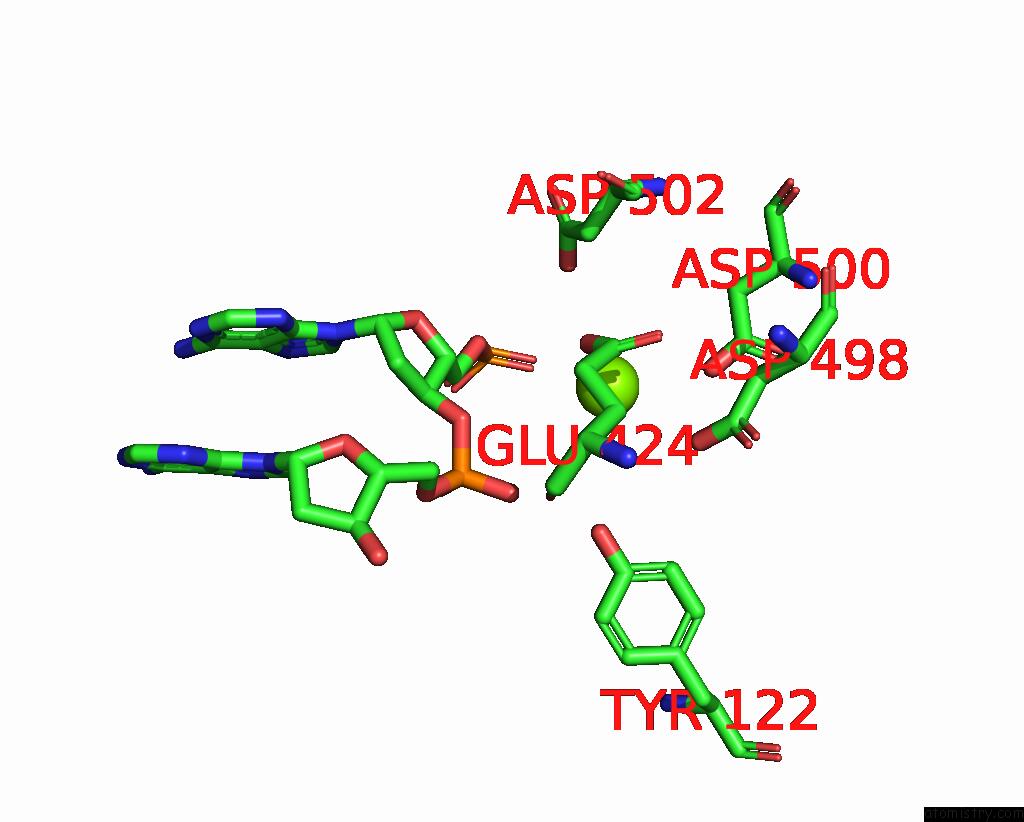
Mono view
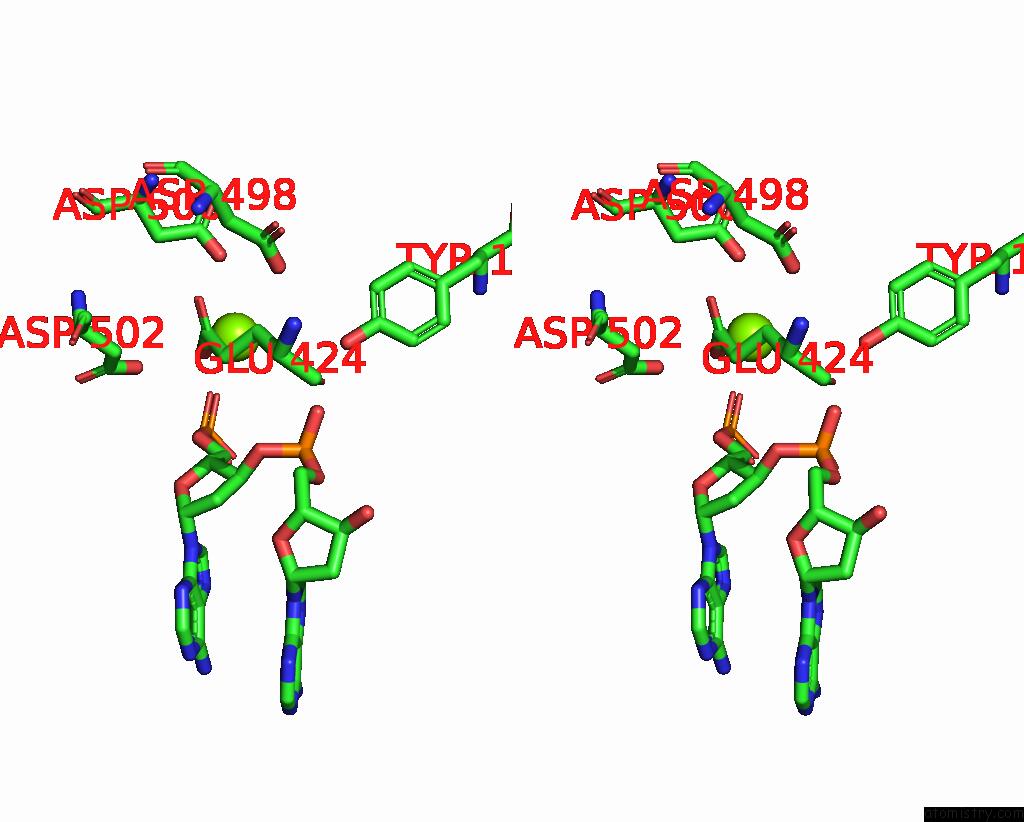
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 1 of E.Coli Dna Gyrase in Complex with 217 Bp Substrate Dna and Lei-800 within 5.0Å range:
|
Magnesium binding site 2 out of 2 in 8qqi
Go back to
Magnesium binding site 2 out
of 2 in the E.Coli Dna Gyrase in Complex with 217 Bp Substrate Dna and Lei-800
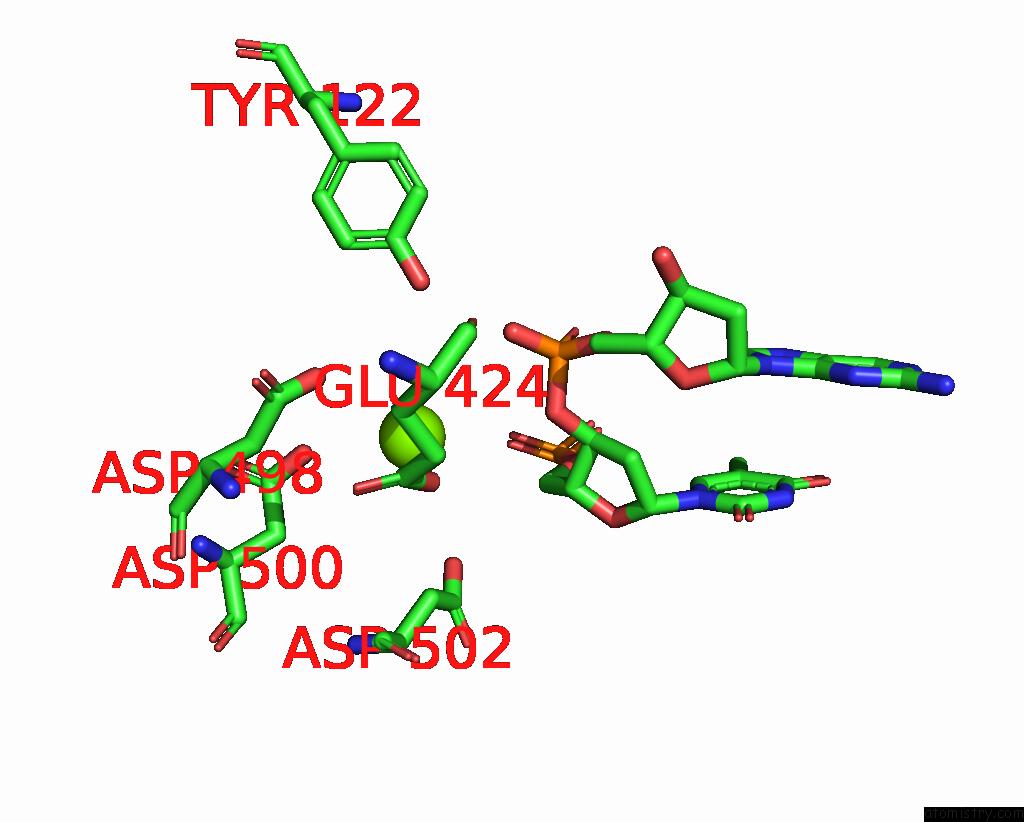
Mono view
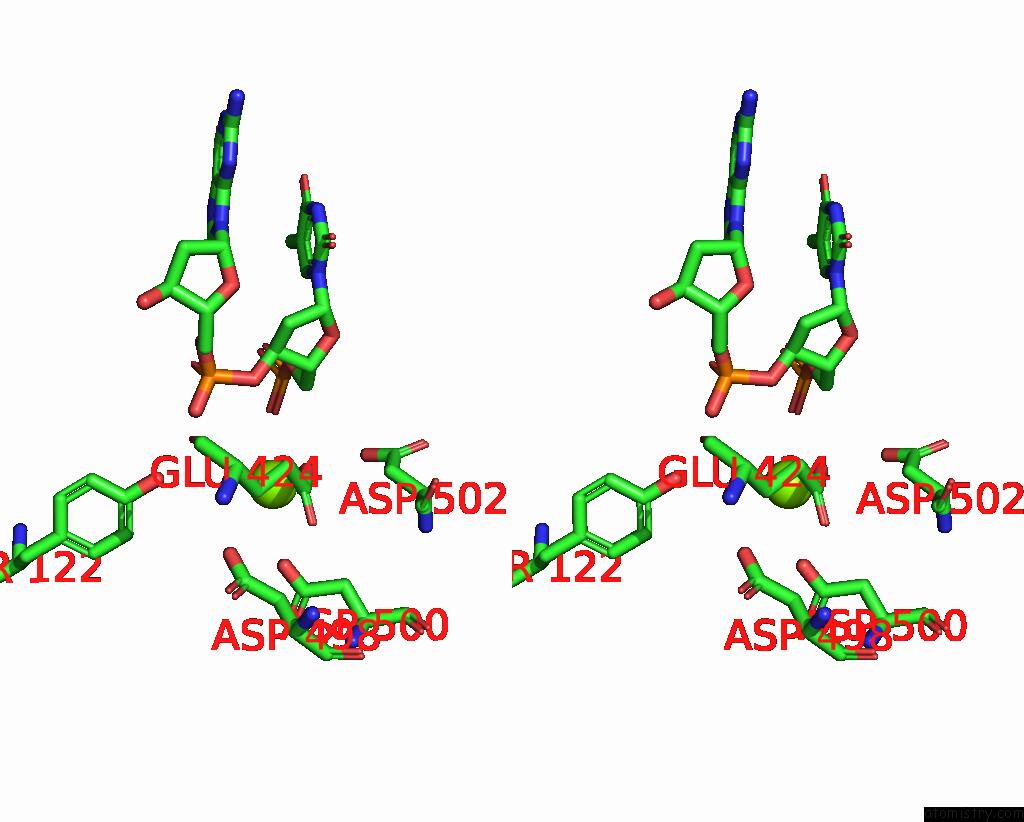
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 2 of E.Coli Dna Gyrase in Complex with 217 Bp Substrate Dna and Lei-800 within 5.0Å range:
|
Reference:
D.Ghilarov,
N.I.Martin,
M.Van Der Stelt.
Discovery of Isoquinoline Sulfonamides As Gyrase Inhibitors Active Against Fluoroquinolone-Resistant Bacteria To Be Published.
Page generated: Fri Aug 15 13:13:58 2025
Last articles
Mg in 8S91Mg in 8S8X
Mg in 8S8K
Mg in 8S8G
Mg in 8S8D
Mg in 8S8E
Mg in 8S8F
Mg in 8S1P
Mg in 8RW1
Mg in 8S87