Magnesium »
PDB 8rkd-8ru2 »
8rqu »
Magnesium in PDB 8rqu: Structure of TEM1 Beta-Lactamase Variant 70.A
Enzymatic activity of Structure of TEM1 Beta-Lactamase Variant 70.A
All present enzymatic activity of Structure of TEM1 Beta-Lactamase Variant 70.A:
3.5.2.6;
3.5.2.6;
Protein crystallography data
The structure of Structure of TEM1 Beta-Lactamase Variant 70.A, PDB code: 8rqu
was solved by
E.Napie,
B.F.Fram,
N.P.Gauthier,
C.Sander,
A.R.Khan,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 41.90 / 2.90 |
| Space group | P 43 |
| Cell size a, b, c (Å), α, β, γ (°) | 114.618, 114.618, 48.882, 90, 90, 90 |
| R / Rfree (%) | 22 / 26.6 |
Magnesium Binding Sites:
The binding sites of Magnesium atom in the Structure of TEM1 Beta-Lactamase Variant 70.A
(pdb code 8rqu). This binding sites where shown within
5.0 Angstroms radius around Magnesium atom.
In total only one binding site of Magnesium was determined in the Structure of TEM1 Beta-Lactamase Variant 70.A, PDB code: 8rqu:
In total only one binding site of Magnesium was determined in the Structure of TEM1 Beta-Lactamase Variant 70.A, PDB code: 8rqu:
Magnesium binding site 1 out of 1 in 8rqu
Go back to
Magnesium binding site 1 out
of 1 in the Structure of TEM1 Beta-Lactamase Variant 70.A
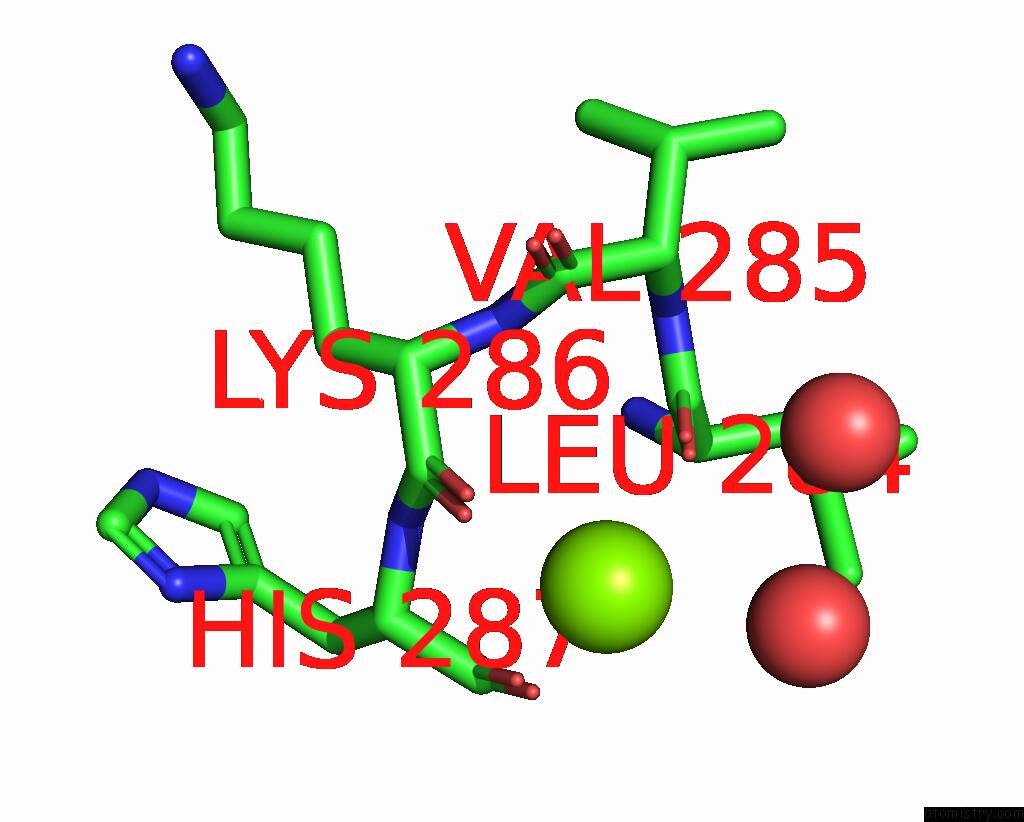
Mono view
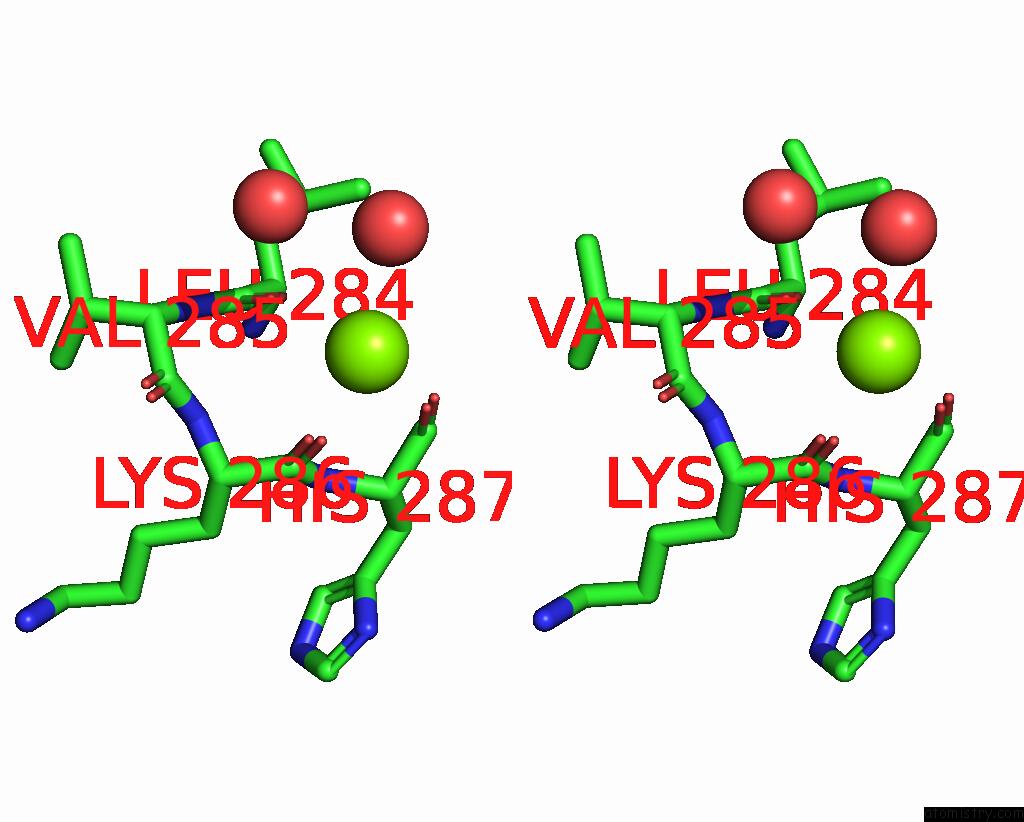
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 1 of Structure of TEM1 Beta-Lactamase Variant 70.A within 5.0Å range:
|
Reference:
B.Fram,
I.Truebridge,
Y.Su,
A.J.Riesselman,
J.B.Ingraham,
A.Passera,
E.Napier,
N.N.Thadani,
S.Lim,
K.Roberts,
G.Kaur,
M.Stiffler,
D.S.Marks,
C.D.Bahl,
A.R.Khan,
C.Sander,
N.P.Gauthier.
Simultaneous Enhancement of Multiple Functional Properties Using Evolution-Informed Protein Design. Biorxiv 2023.
ISSN: ISSN 2692-8205
PubMed: 37214973
DOI: 10.1101/2023.05.09.539914
Page generated: Fri Aug 15 14:07:31 2025
ISSN: ISSN 2692-8205
PubMed: 37214973
DOI: 10.1101/2023.05.09.539914
Last articles
Mg in 9FC1Mg in 9FBZ
Mg in 9FC0
Mg in 9FBN
Mg in 9FBJ
Mg in 9FB9
Mg in 9F6C
Mg in 9FB8
Mg in 9F6D
Mg in 9FB7