Magnesium »
PDB 8tef-8tlk »
8thd »
Magnesium in PDB 8thd: Structure of the Saccharomyces Cerevisiae Clamp Unloader ELG1-Rfc Bound to Pcna
Magnesium Binding Sites:
The binding sites of Magnesium atom in the Structure of the Saccharomyces Cerevisiae Clamp Unloader ELG1-Rfc Bound to Pcna
(pdb code 8thd). This binding sites where shown within
5.0 Angstroms radius around Magnesium atom.
In total 3 binding sites of Magnesium where determined in the Structure of the Saccharomyces Cerevisiae Clamp Unloader ELG1-Rfc Bound to Pcna, PDB code: 8thd:
Jump to Magnesium binding site number: 1; 2; 3;
In total 3 binding sites of Magnesium where determined in the Structure of the Saccharomyces Cerevisiae Clamp Unloader ELG1-Rfc Bound to Pcna, PDB code: 8thd:
Jump to Magnesium binding site number: 1; 2; 3;
Magnesium binding site 1 out of 3 in 8thd
Go back to
Magnesium binding site 1 out
of 3 in the Structure of the Saccharomyces Cerevisiae Clamp Unloader ELG1-Rfc Bound to Pcna
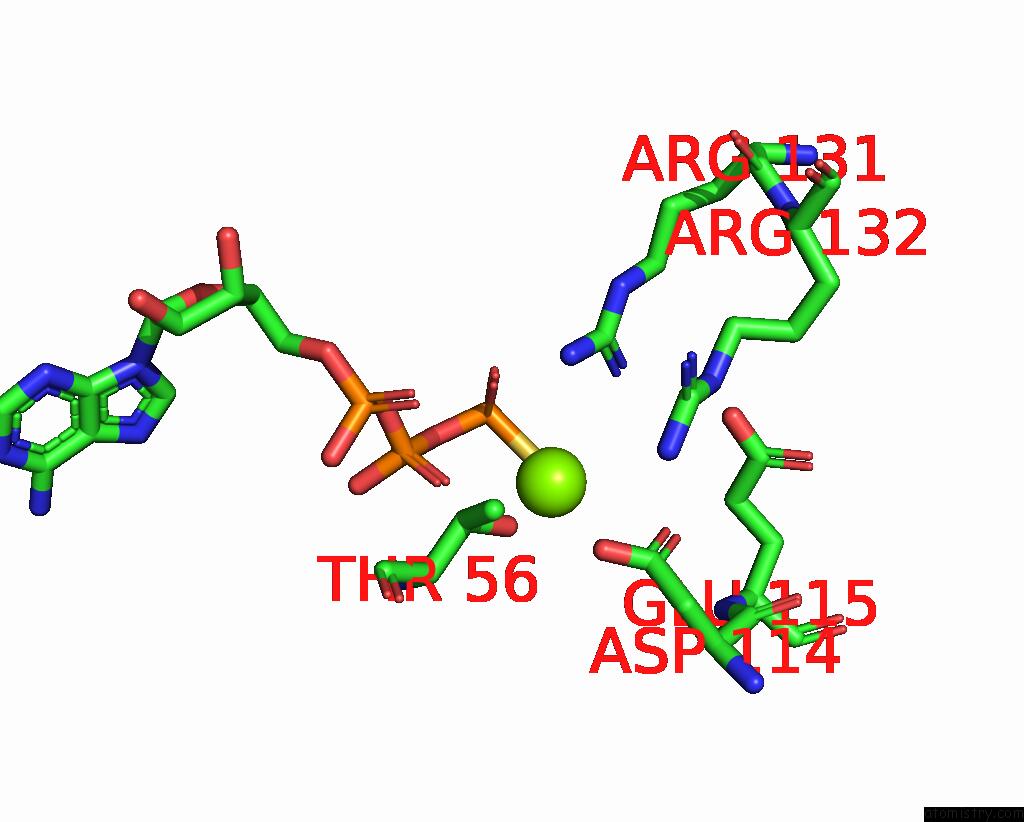
Mono view
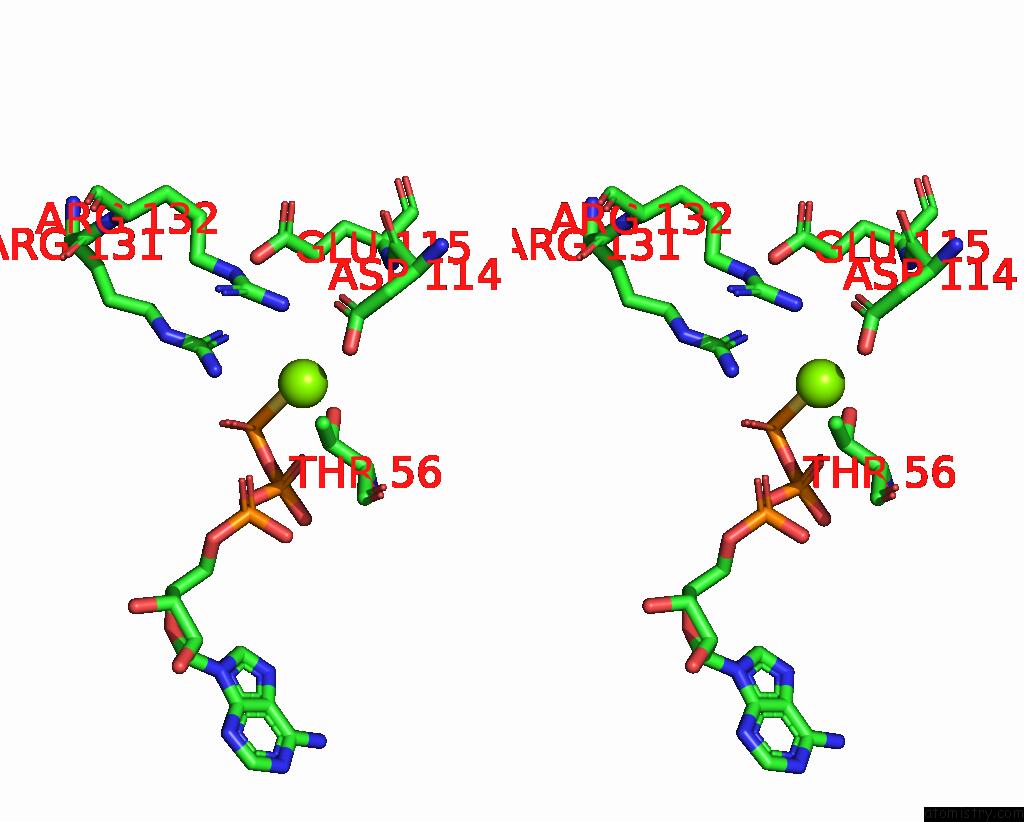
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 1 of Structure of the Saccharomyces Cerevisiae Clamp Unloader ELG1-Rfc Bound to Pcna within 5.0Å range:
|
Magnesium binding site 2 out of 3 in 8thd
Go back to
Magnesium binding site 2 out
of 3 in the Structure of the Saccharomyces Cerevisiae Clamp Unloader ELG1-Rfc Bound to Pcna
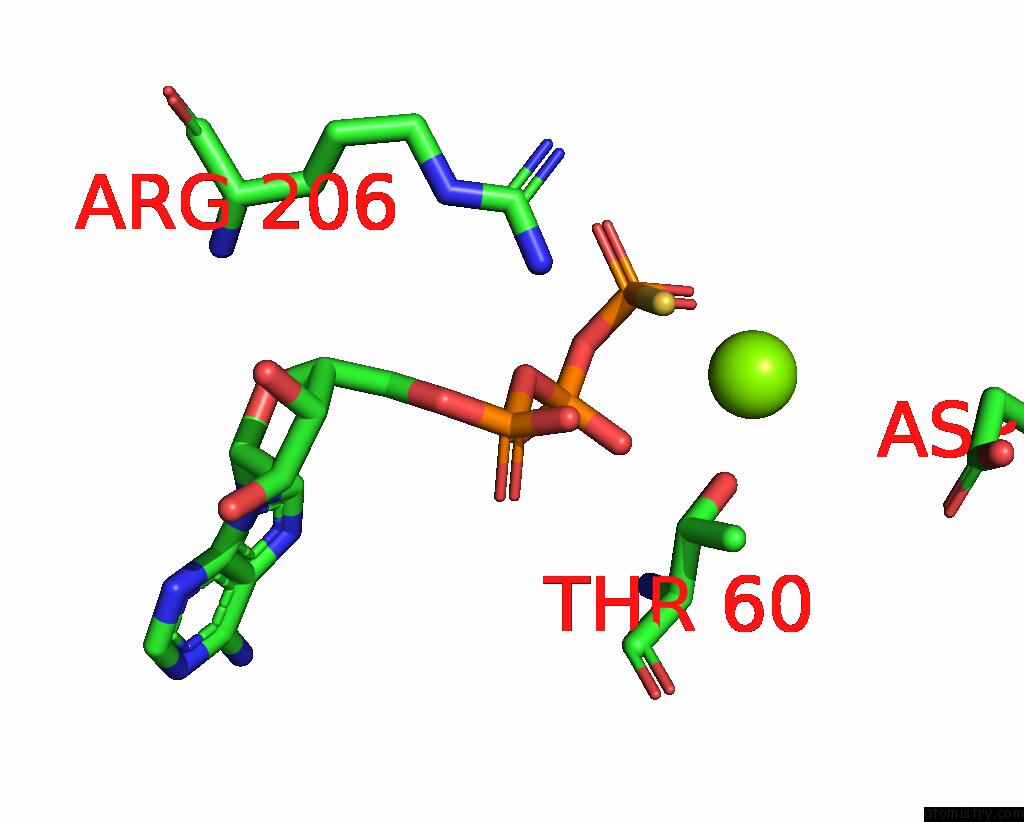
Mono view
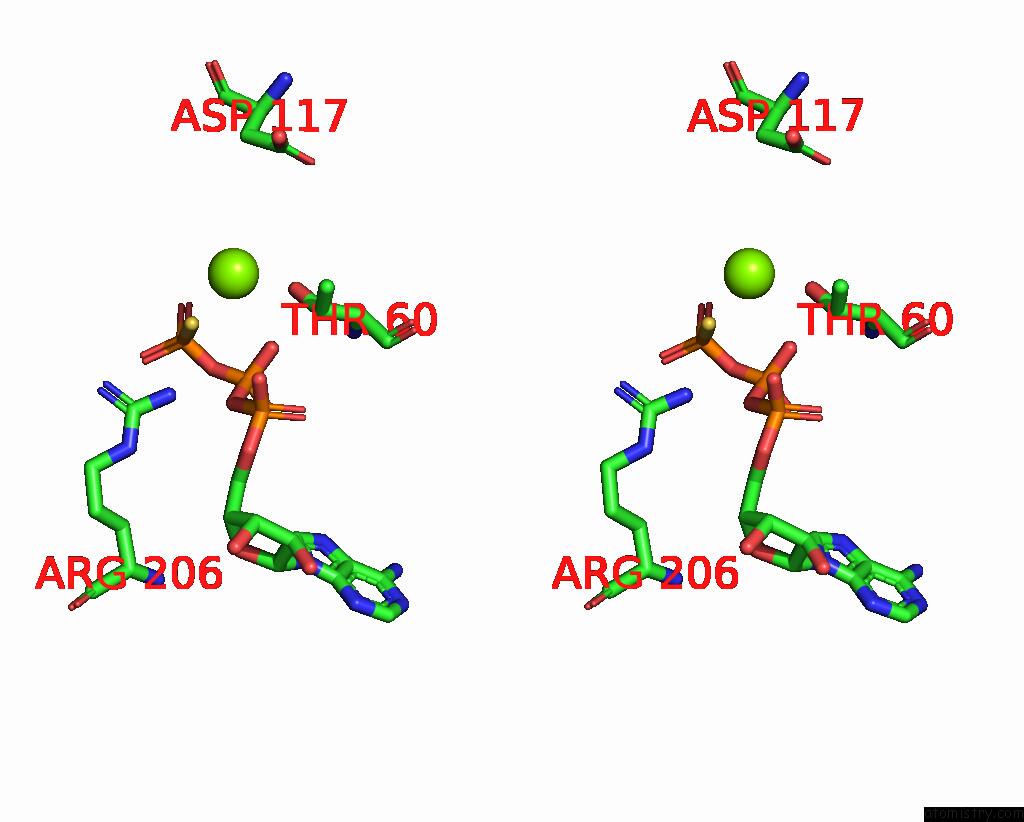
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 2 of Structure of the Saccharomyces Cerevisiae Clamp Unloader ELG1-Rfc Bound to Pcna within 5.0Å range:
|
Magnesium binding site 3 out of 3 in 8thd
Go back to
Magnesium binding site 3 out
of 3 in the Structure of the Saccharomyces Cerevisiae Clamp Unloader ELG1-Rfc Bound to Pcna
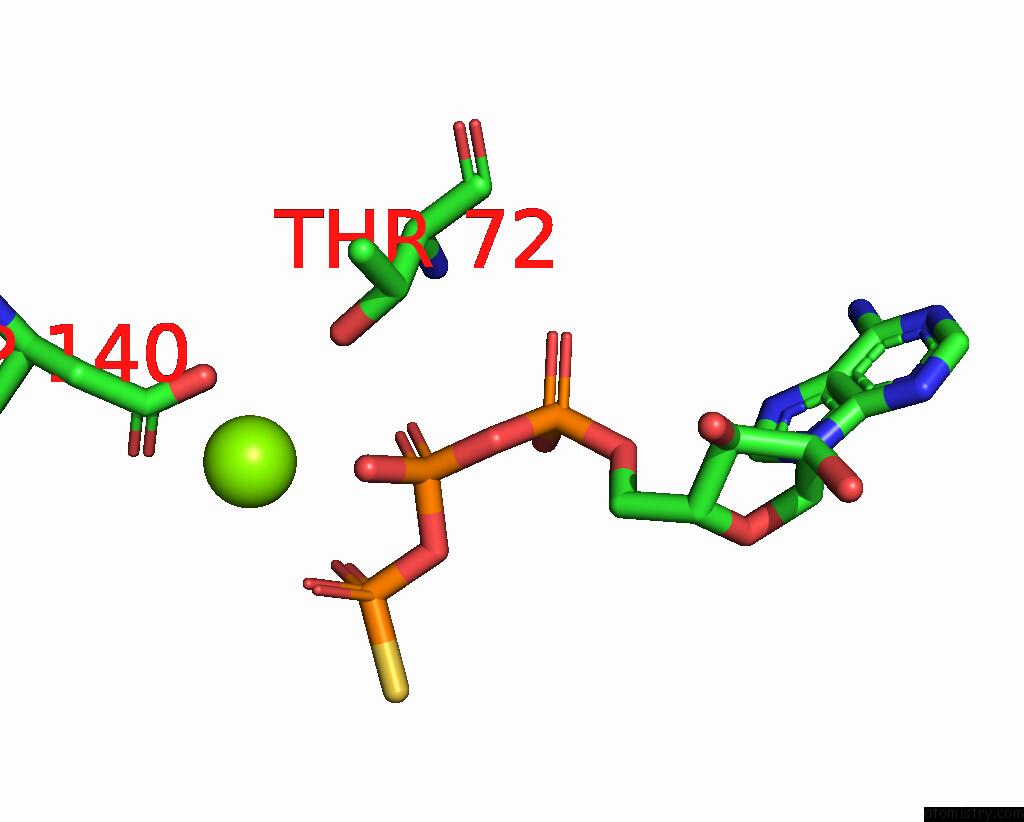
Mono view
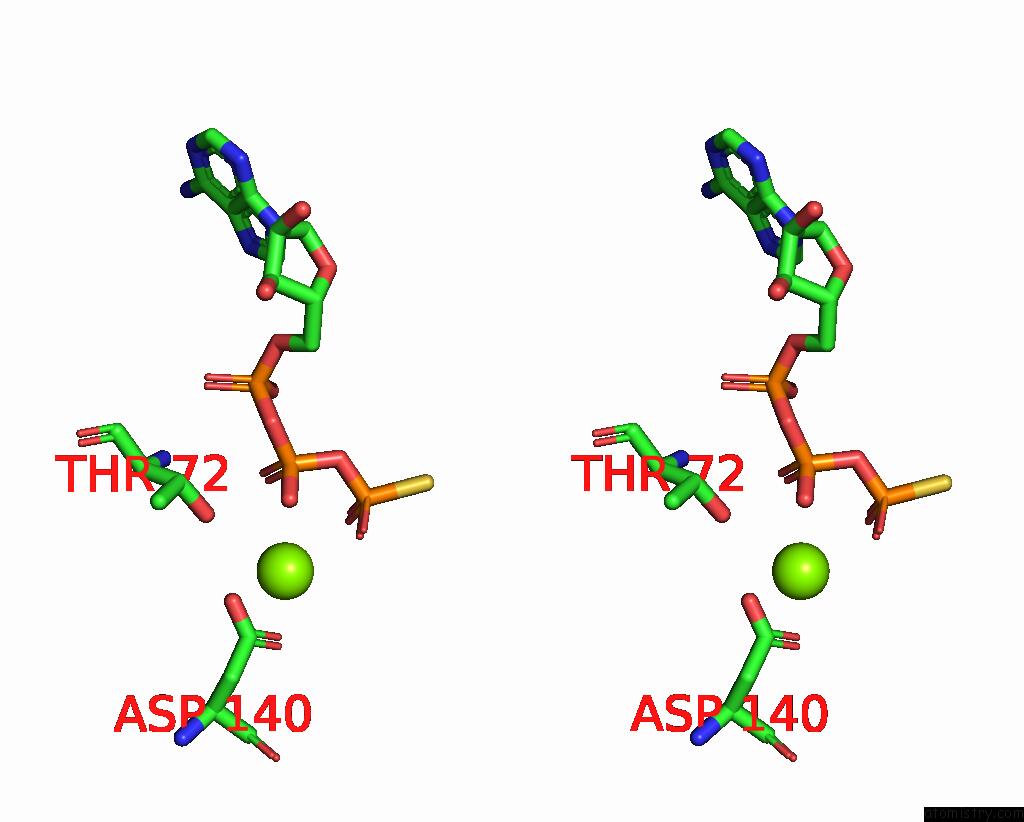
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 3 of Structure of the Saccharomyces Cerevisiae Clamp Unloader ELG1-Rfc Bound to Pcna within 5.0Å range:
|
Reference:
F.Zheng,
Y.N.Yao,
R.Georgescu,
M.E.O'donnell,
H.Li.
Structure of the Pcna Unloader ELG1-Rfc Sci Adv 2024.
ISSN: ESSN 2375-2548
DOI: 10.1126/SCIADV.ADL1739
Page generated: Fri Aug 15 16:21:33 2025
ISSN: ESSN 2375-2548
DOI: 10.1126/SCIADV.ADL1739
Last articles
Mg in 8WKGMg in 8WKF
Mg in 8WIM
Mg in 8WIL
Mg in 8WH2
Mg in 8WH0
Mg in 8WGY
Mg in 8WDV
Mg in 8WGO
Mg in 8WDU