Magnesium »
PDB 8u3b-8ucw »
8u72 »
Magnesium in PDB 8u72: Cryo-Em Structure of the Sparta Oligomer with Guide Rna and Target Dna
Magnesium Binding Sites:
The binding sites of Magnesium atom in the Cryo-Em Structure of the Sparta Oligomer with Guide Rna and Target Dna
(pdb code 8u72). This binding sites where shown within
5.0 Angstroms radius around Magnesium atom.
In total 4 binding sites of Magnesium where determined in the Cryo-Em Structure of the Sparta Oligomer with Guide Rna and Target Dna, PDB code: 8u72:
Jump to Magnesium binding site number: 1; 2; 3; 4;
In total 4 binding sites of Magnesium where determined in the Cryo-Em Structure of the Sparta Oligomer with Guide Rna and Target Dna, PDB code: 8u72:
Jump to Magnesium binding site number: 1; 2; 3; 4;
Magnesium binding site 1 out of 4 in 8u72
Go back to
Magnesium binding site 1 out
of 4 in the Cryo-Em Structure of the Sparta Oligomer with Guide Rna and Target Dna
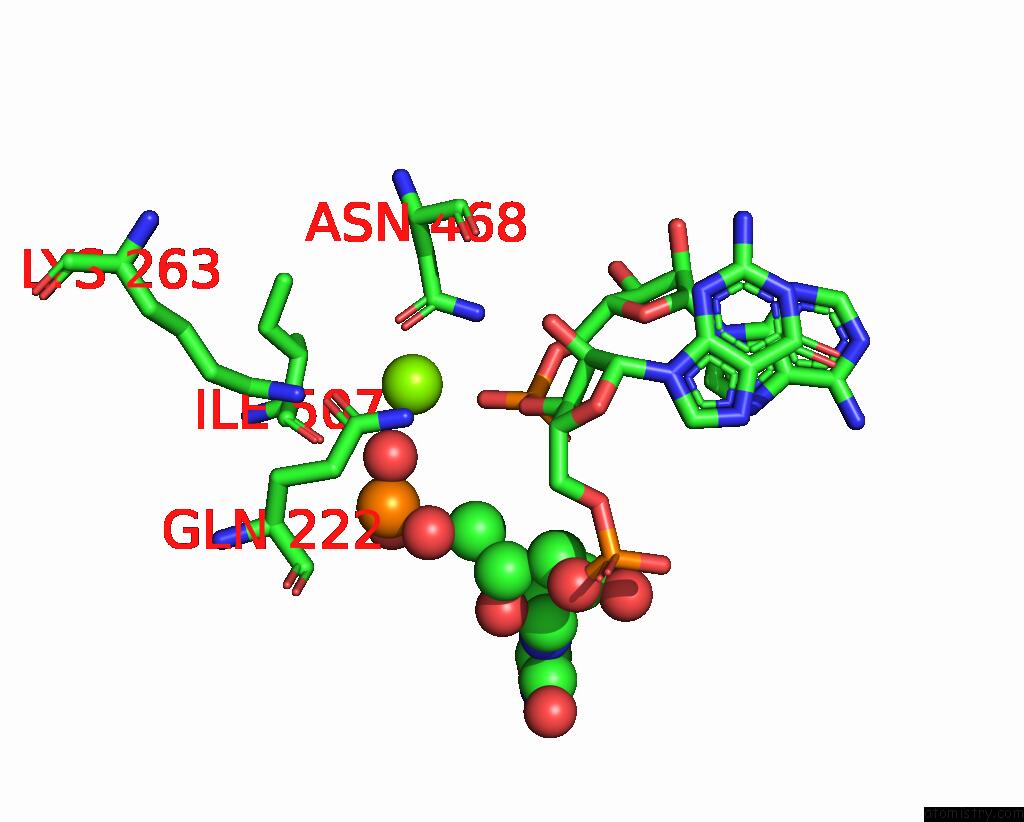
Mono view
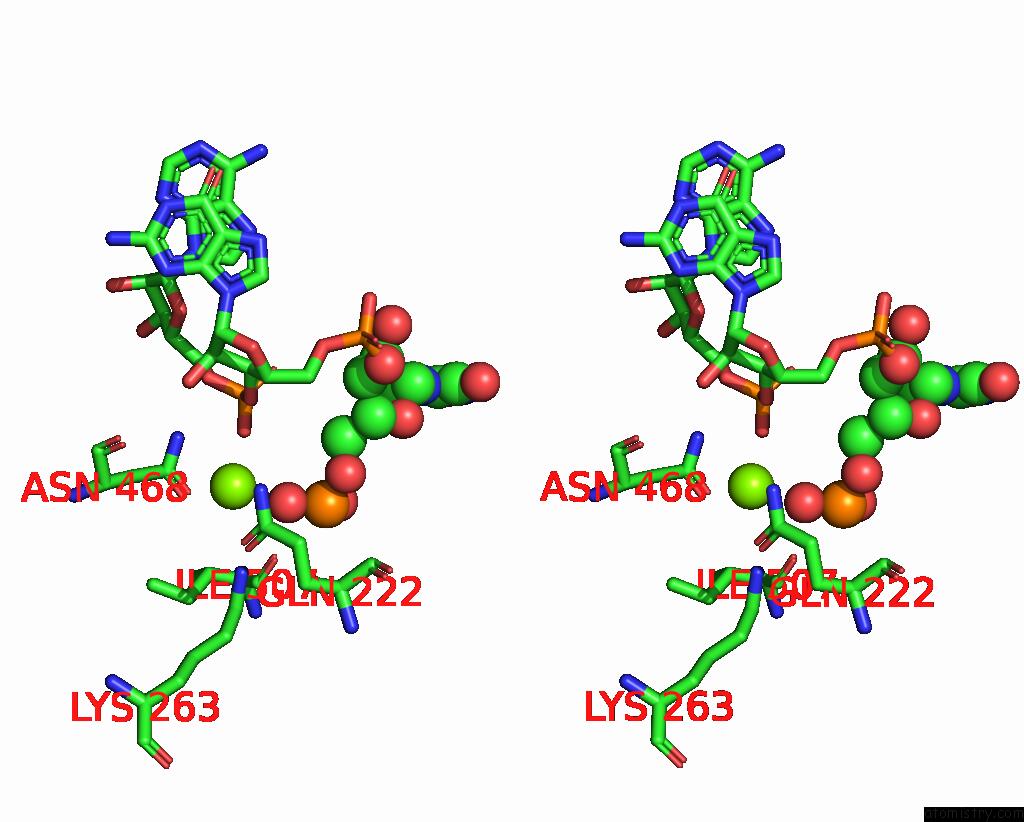
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 1 of Cryo-Em Structure of the Sparta Oligomer with Guide Rna and Target Dna within 5.0Å range:
|
Magnesium binding site 2 out of 4 in 8u72
Go back to
Magnesium binding site 2 out
of 4 in the Cryo-Em Structure of the Sparta Oligomer with Guide Rna and Target Dna
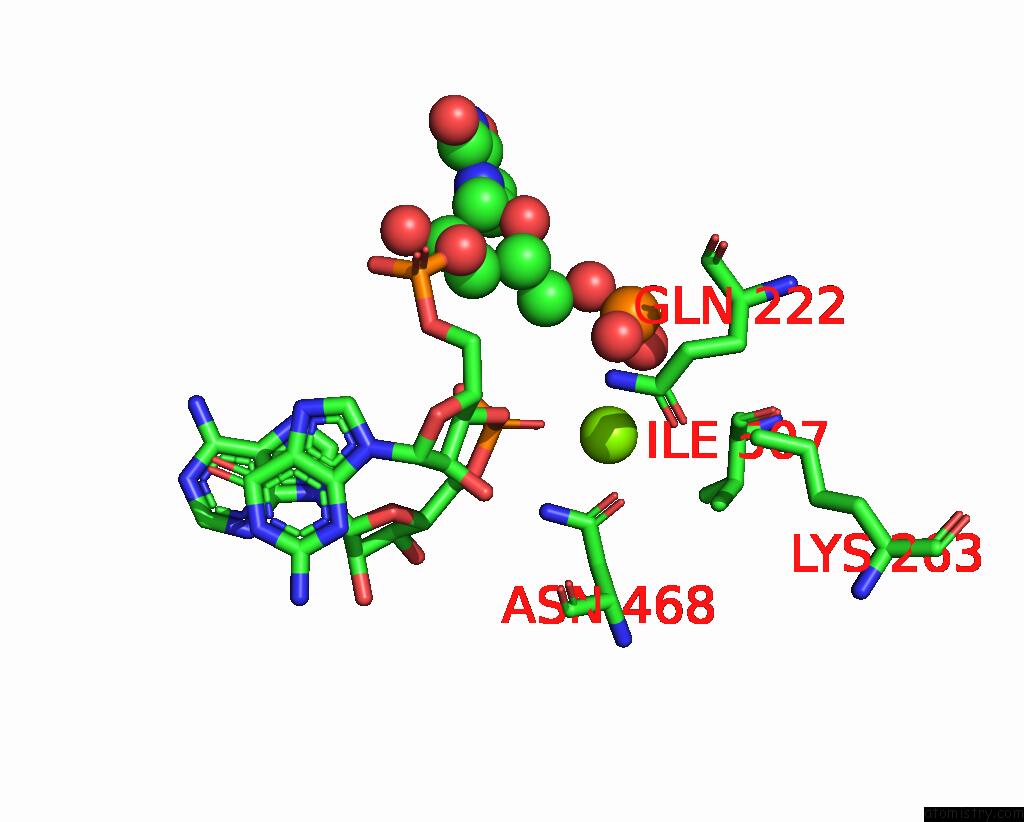
Mono view
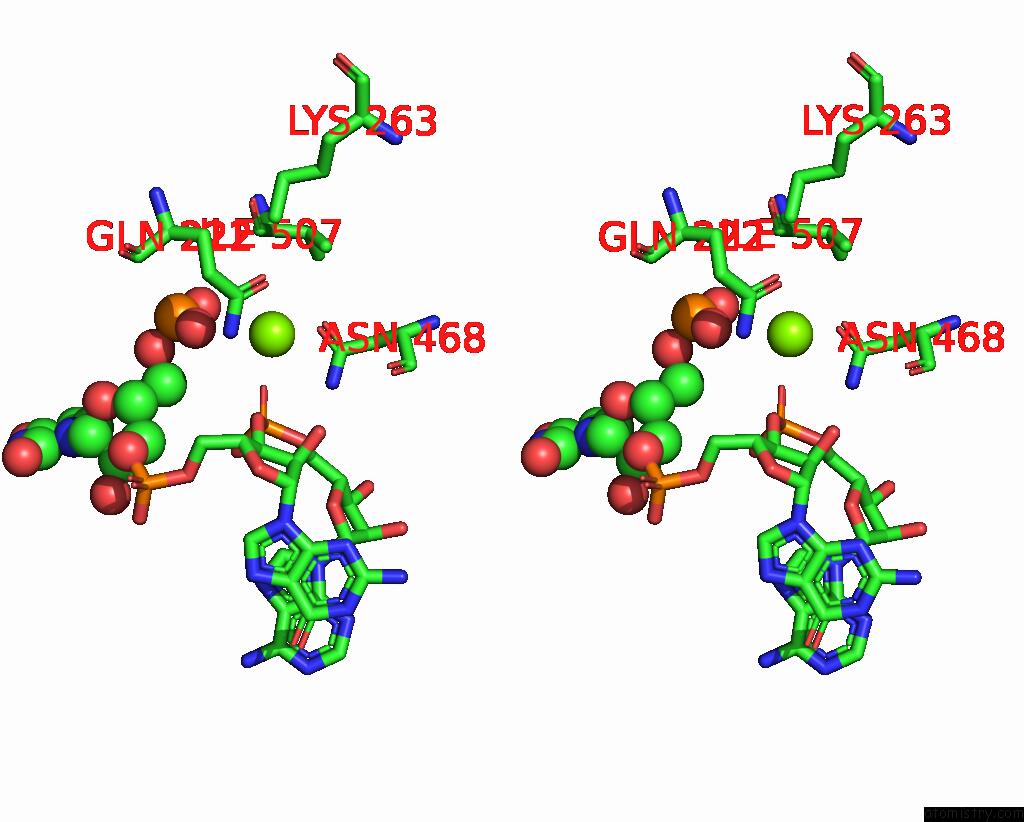
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 2 of Cryo-Em Structure of the Sparta Oligomer with Guide Rna and Target Dna within 5.0Å range:
|
Magnesium binding site 3 out of 4 in 8u72
Go back to
Magnesium binding site 3 out
of 4 in the Cryo-Em Structure of the Sparta Oligomer with Guide Rna and Target Dna
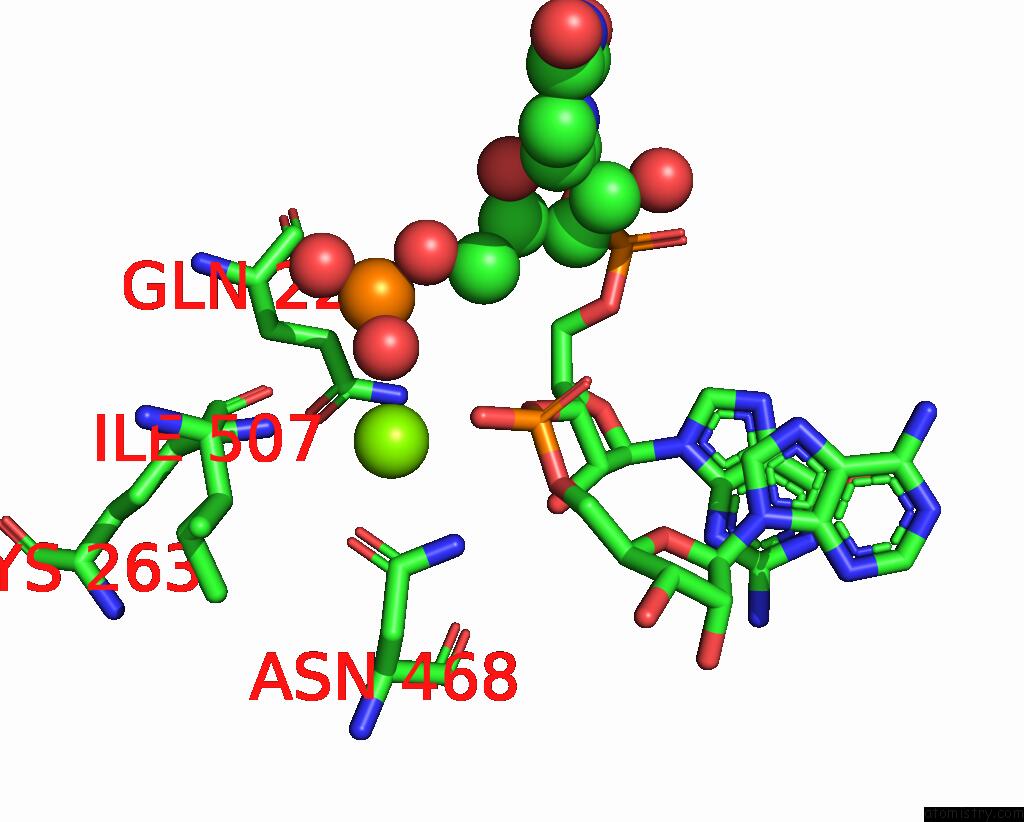
Mono view
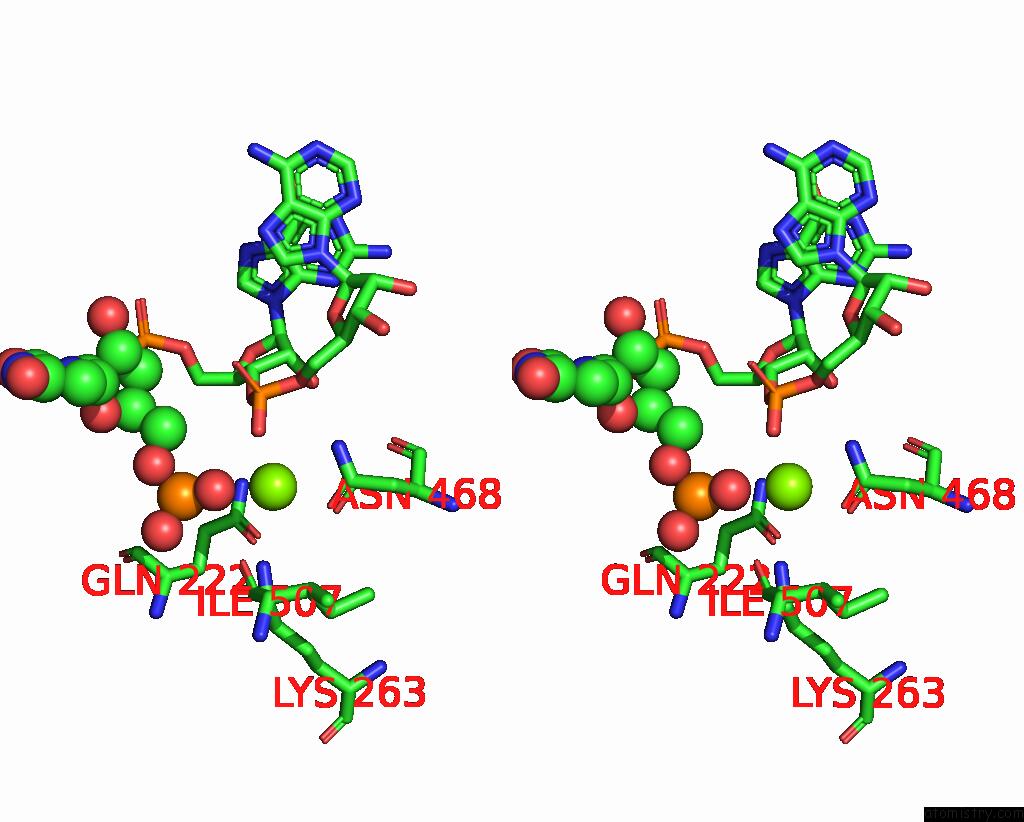
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 3 of Cryo-Em Structure of the Sparta Oligomer with Guide Rna and Target Dna within 5.0Å range:
|
Magnesium binding site 4 out of 4 in 8u72
Go back to
Magnesium binding site 4 out
of 4 in the Cryo-Em Structure of the Sparta Oligomer with Guide Rna and Target Dna
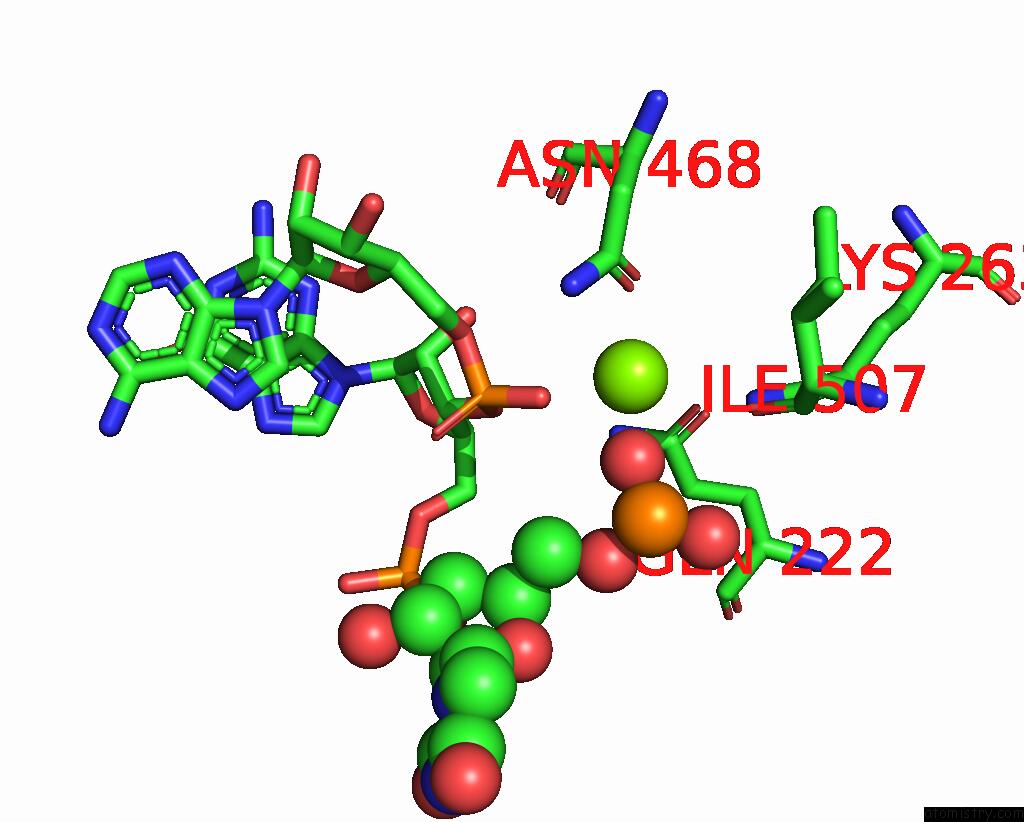
Mono view
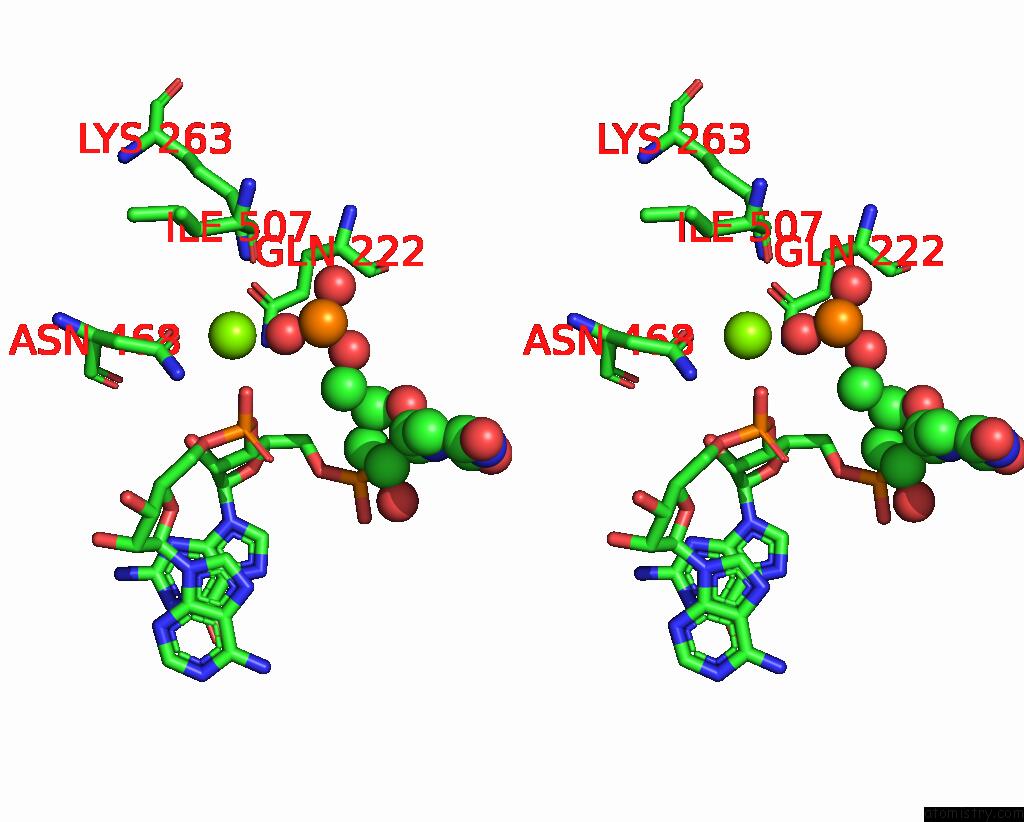
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 4 of Cryo-Em Structure of the Sparta Oligomer with Guide Rna and Target Dna within 5.0Å range:
|
Reference:
J.Kottur,
R.Malik,
A.K.Aggarwal.
Nucleic Acid Mediated Activation of Prokaryotic Argonaute Immune System Nat Commun 2024.
ISSN: ESSN 2041-1723
Page generated: Fri Aug 15 16:48:57 2025
ISSN: ESSN 2041-1723
Last articles
Mg in 8WGHMg in 8WMO
Mg in 8WMN
Mg in 8WMM
Mg in 8WEY
Mg in 8WKG
Mg in 8WKF
Mg in 8WIM
Mg in 8WIL
Mg in 8WH2