Magnesium »
PDB 8v4k-8vbg »
8v6v »
Magnesium in PDB 8v6v: Cryo-Em Structure of Doubly-Bound SNF2H-Nucleosome Complex
Magnesium Binding Sites:
The binding sites of Magnesium atom in the Cryo-Em Structure of Doubly-Bound SNF2H-Nucleosome Complex
(pdb code 8v6v). This binding sites where shown within
5.0 Angstroms radius around Magnesium atom.
In total 2 binding sites of Magnesium where determined in the Cryo-Em Structure of Doubly-Bound SNF2H-Nucleosome Complex, PDB code: 8v6v:
Jump to Magnesium binding site number: 1; 2;
In total 2 binding sites of Magnesium where determined in the Cryo-Em Structure of Doubly-Bound SNF2H-Nucleosome Complex, PDB code: 8v6v:
Jump to Magnesium binding site number: 1; 2;
Magnesium binding site 1 out of 2 in 8v6v
Go back to
Magnesium binding site 1 out
of 2 in the Cryo-Em Structure of Doubly-Bound SNF2H-Nucleosome Complex
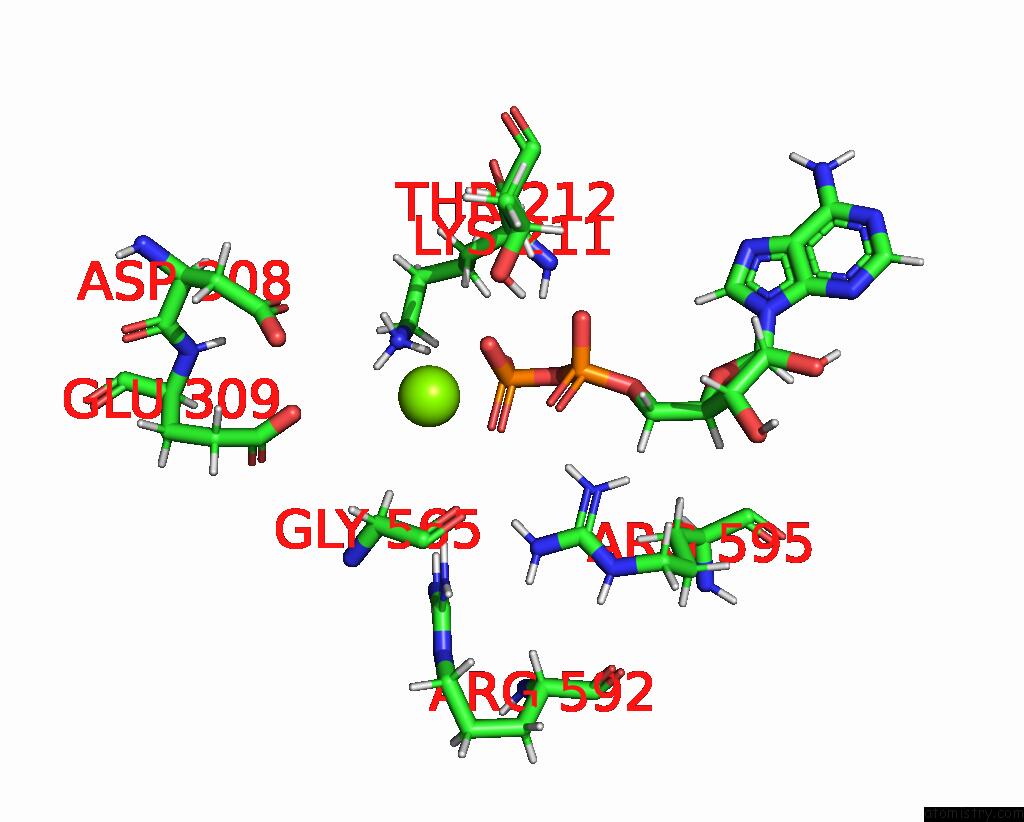
Mono view
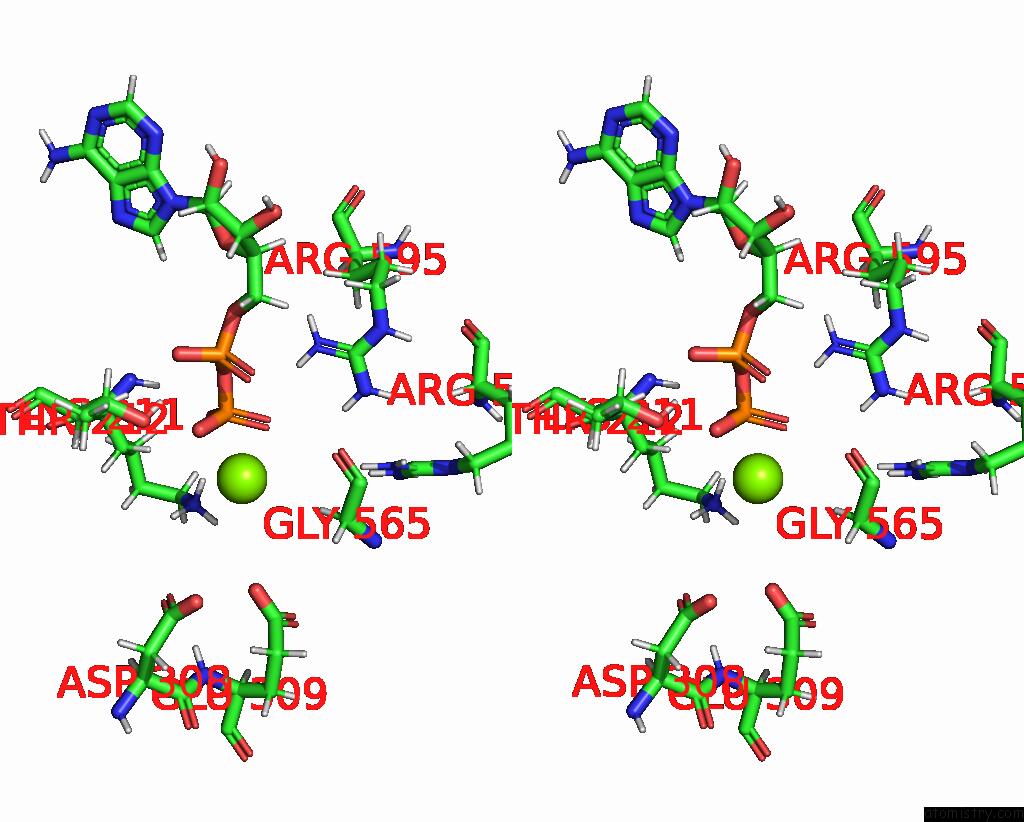
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 1 of Cryo-Em Structure of Doubly-Bound SNF2H-Nucleosome Complex within 5.0Å range:
|
Magnesium binding site 2 out of 2 in 8v6v
Go back to
Magnesium binding site 2 out
of 2 in the Cryo-Em Structure of Doubly-Bound SNF2H-Nucleosome Complex
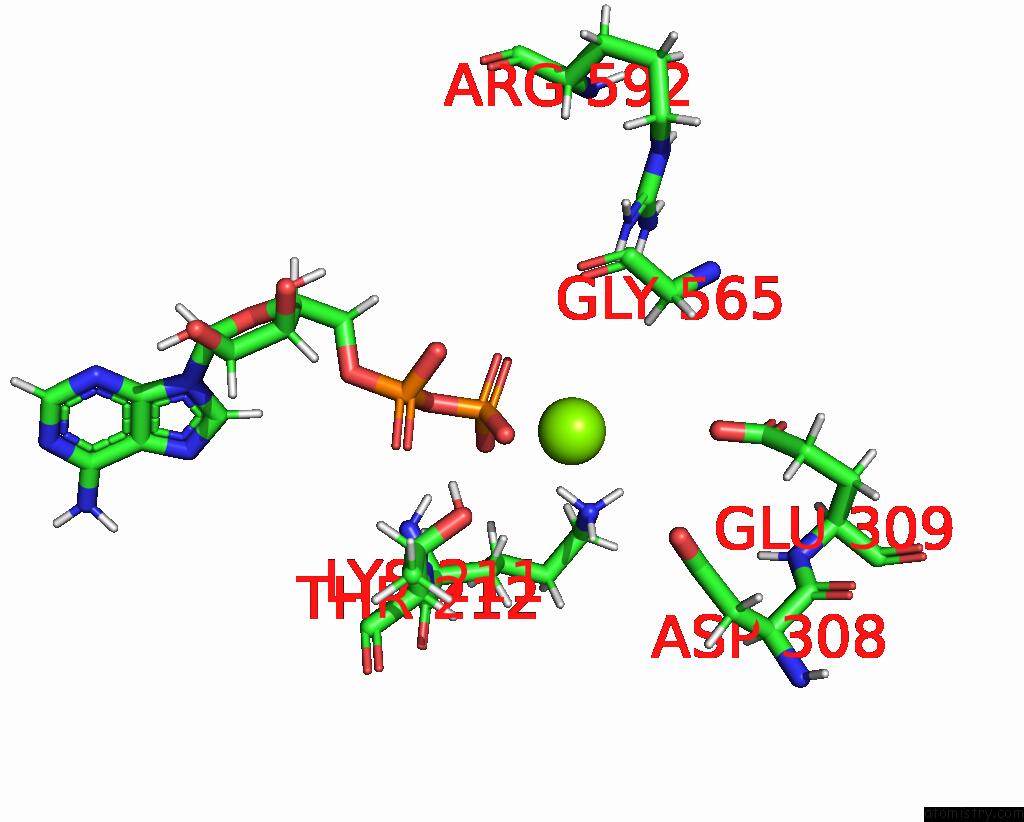
Mono view
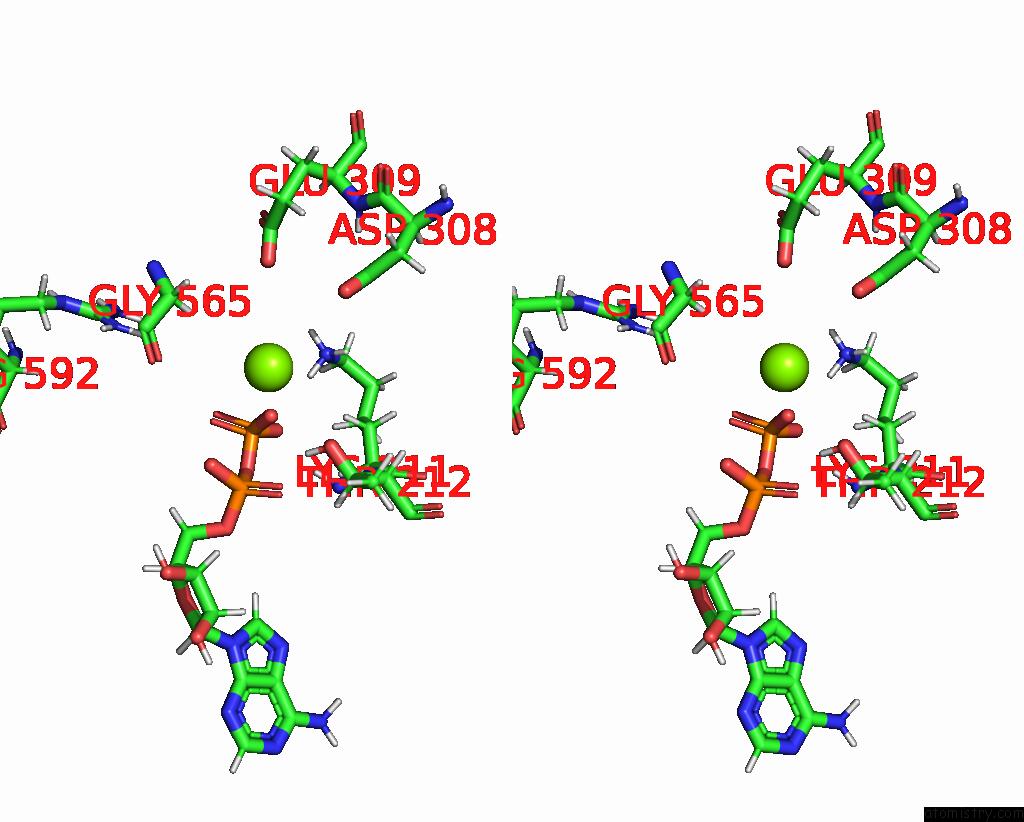
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 2 of Cryo-Em Structure of Doubly-Bound SNF2H-Nucleosome Complex within 5.0Å range:
|
Reference:
U.S.Chio,
E.Palovcak,
A.A.A.Smith,
H.Autzen,
E.N.Munoz,
Z.Yu,
F.Wang,
D.A.Agard,
J.P.Armache,
G.J.Narlikar,
Y.Cheng.
Functionalized Graphene-Oxide Grids Enable High-Resolution Cryo-Em Structures of the SNF2H-Nucleosome Complex Without Crosslinking. Nat Commun V. 15 2225 2024.
ISSN: ESSN 2041-1723
PubMed: 38472177
DOI: 10.1038/S41467-024-46178-Y
Page generated: Fri Aug 15 17:30:21 2025
ISSN: ESSN 2041-1723
PubMed: 38472177
DOI: 10.1038/S41467-024-46178-Y
Last articles
Mg in 9BY7Mg in 9BY3
Mg in 9BY2
Mg in 9BY1
Mg in 9BXZ
Mg in 9BY0
Mg in 9BXX
Mg in 9BXT
Mg in 9BXS
Mg in 9BX9