Magnesium »
PDB 8vbi-8vn3 »
8vct »
Magnesium in PDB 8vct: Cyoem Structure of the Tnsc(1-503)-Tnsd(1-318)-Dna Complex in A 6:2:1 Stoichiometry From E. Coli TN7 Bound to Atpgs and Adp
Other elements in 8vct:
The structure of Cyoem Structure of the Tnsc(1-503)-Tnsd(1-318)-Dna Complex in A 6:2:1 Stoichiometry From E. Coli TN7 Bound to Atpgs and Adp also contains other interesting chemical elements:
| Zinc | (Zn) | 2 atoms |
Magnesium Binding Sites:
The binding sites of Magnesium atom in the Cyoem Structure of the Tnsc(1-503)-Tnsd(1-318)-Dna Complex in A 6:2:1 Stoichiometry From E. Coli TN7 Bound to Atpgs and Adp
(pdb code 8vct). This binding sites where shown within
5.0 Angstroms radius around Magnesium atom.
In total 6 binding sites of Magnesium where determined in the Cyoem Structure of the Tnsc(1-503)-Tnsd(1-318)-Dna Complex in A 6:2:1 Stoichiometry From E. Coli TN7 Bound to Atpgs and Adp, PDB code: 8vct:
Jump to Magnesium binding site number: 1; 2; 3; 4; 5; 6;
In total 6 binding sites of Magnesium where determined in the Cyoem Structure of the Tnsc(1-503)-Tnsd(1-318)-Dna Complex in A 6:2:1 Stoichiometry From E. Coli TN7 Bound to Atpgs and Adp, PDB code: 8vct:
Jump to Magnesium binding site number: 1; 2; 3; 4; 5; 6;
Magnesium binding site 1 out of 6 in 8vct
Go back to
Magnesium binding site 1 out
of 6 in the Cyoem Structure of the Tnsc(1-503)-Tnsd(1-318)-Dna Complex in A 6:2:1 Stoichiometry From E. Coli TN7 Bound to Atpgs and Adp
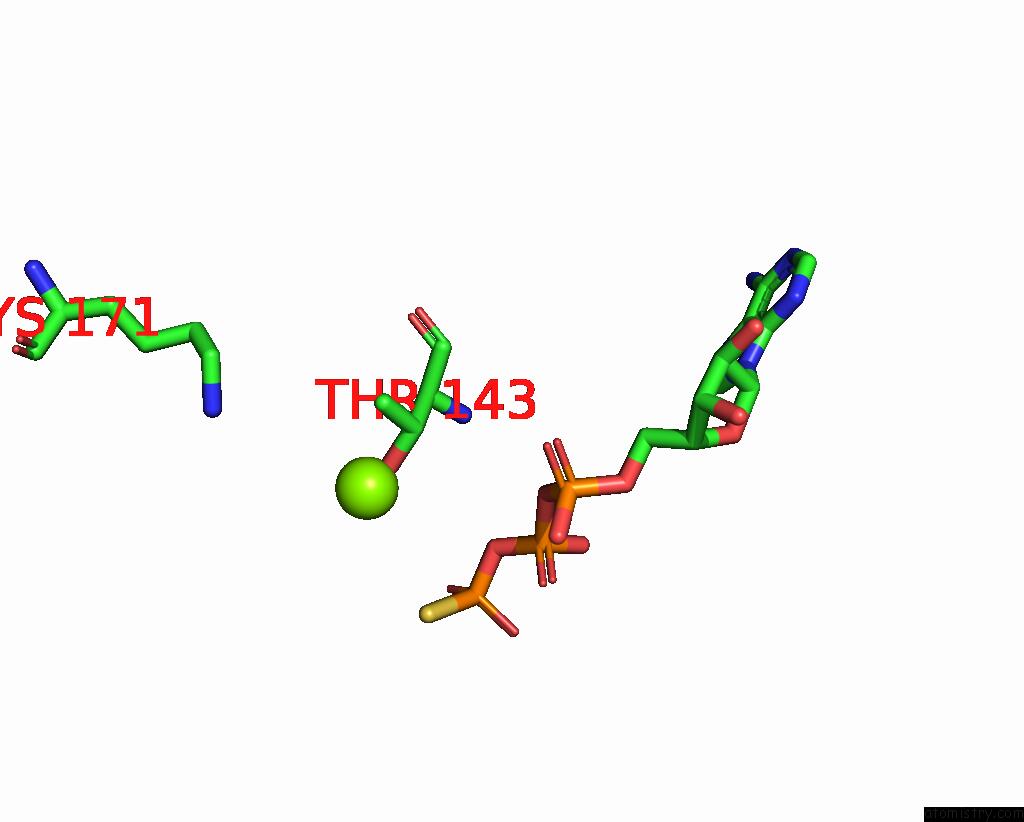
Mono view
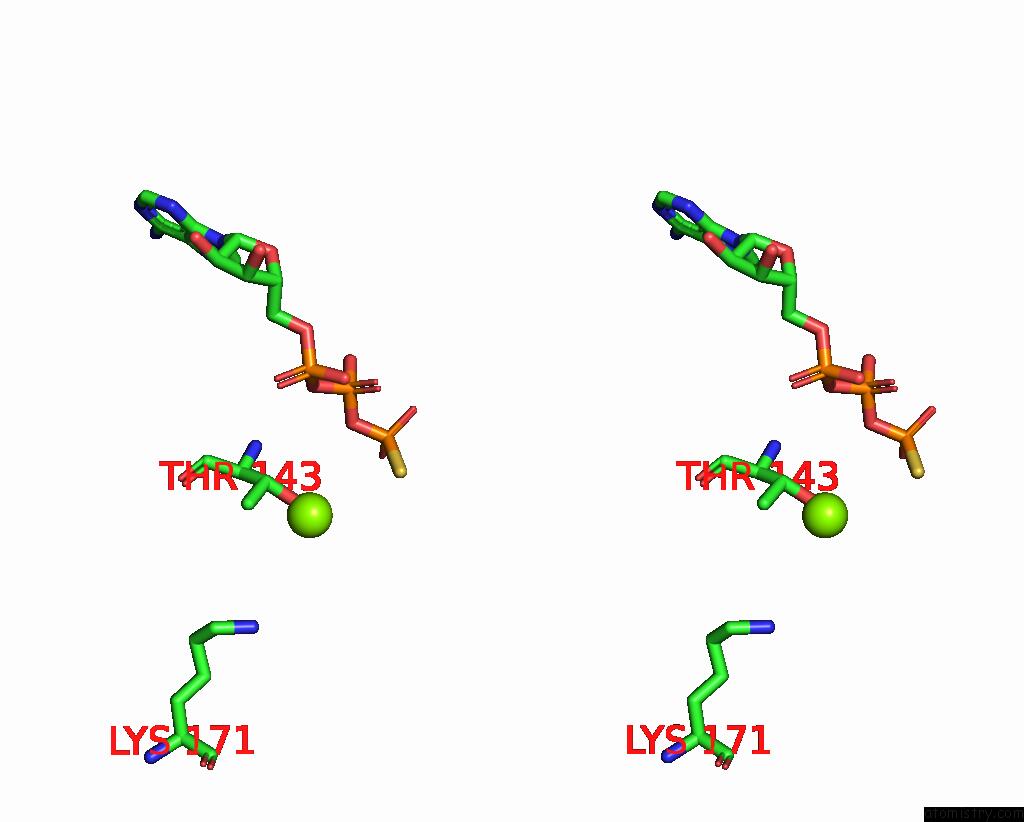
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 1 of Cyoem Structure of the Tnsc(1-503)-Tnsd(1-318)-Dna Complex in A 6:2:1 Stoichiometry From E. Coli TN7 Bound to Atpgs and Adp within 5.0Å range:
|
Magnesium binding site 2 out of 6 in 8vct
Go back to
Magnesium binding site 2 out
of 6 in the Cyoem Structure of the Tnsc(1-503)-Tnsd(1-318)-Dna Complex in A 6:2:1 Stoichiometry From E. Coli TN7 Bound to Atpgs and Adp
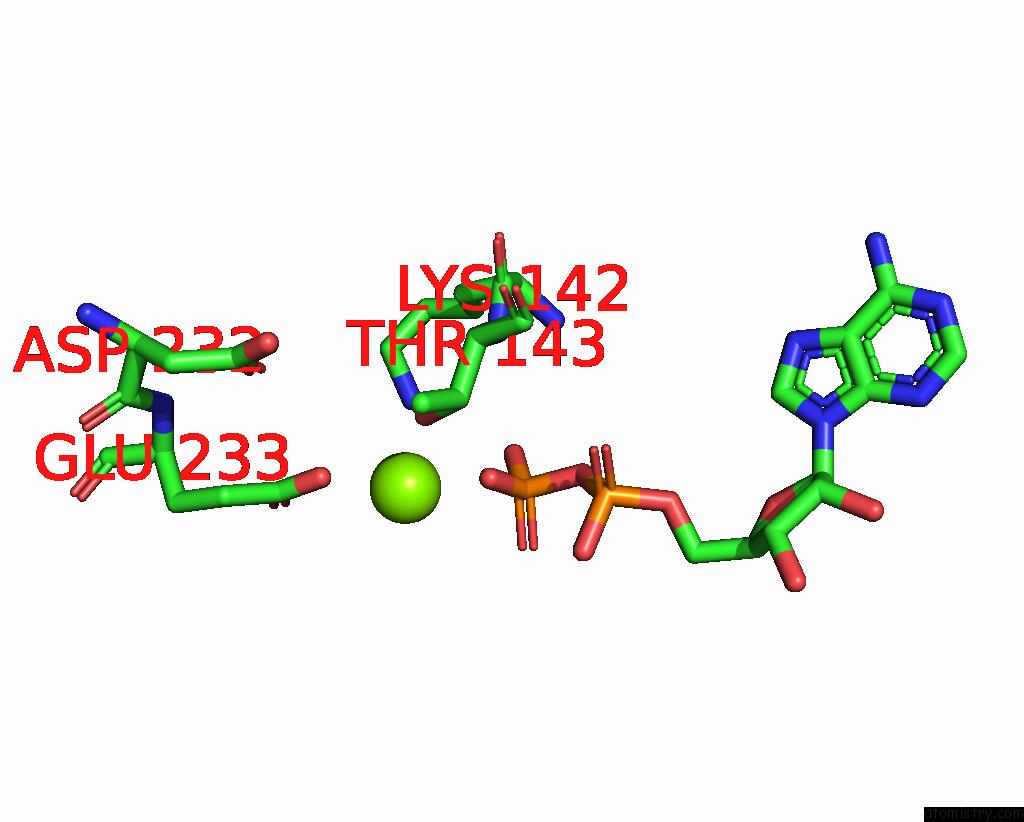
Mono view
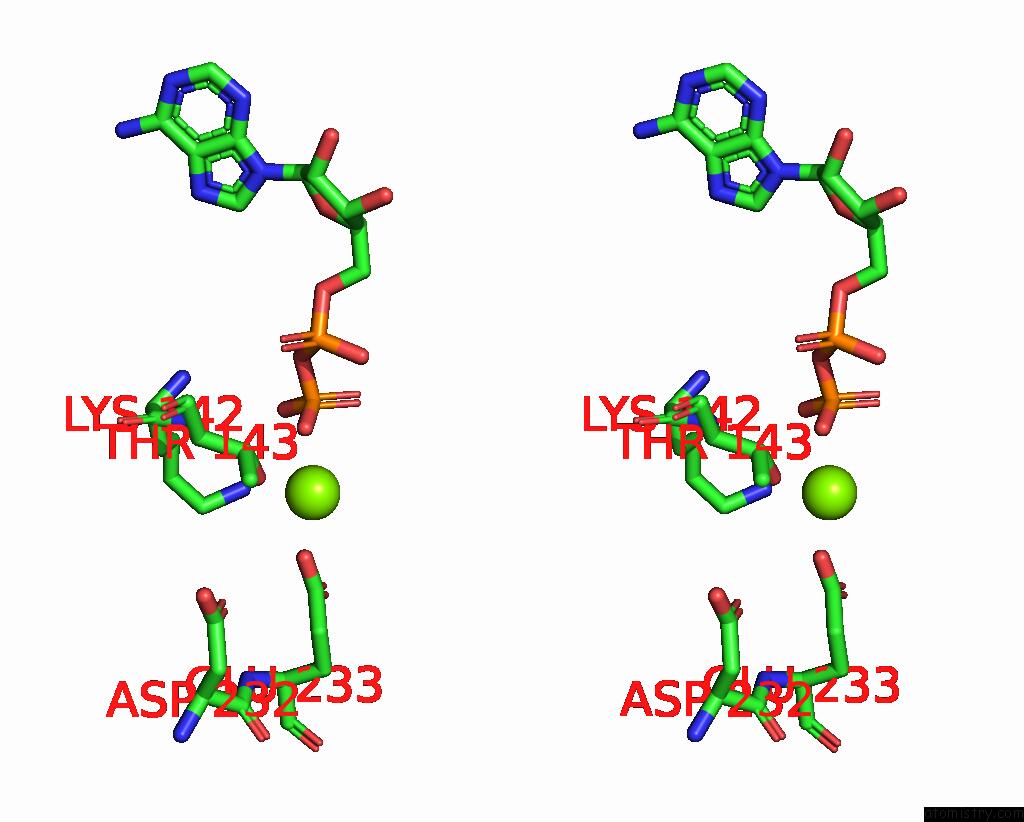
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 2 of Cyoem Structure of the Tnsc(1-503)-Tnsd(1-318)-Dna Complex in A 6:2:1 Stoichiometry From E. Coli TN7 Bound to Atpgs and Adp within 5.0Å range:
|
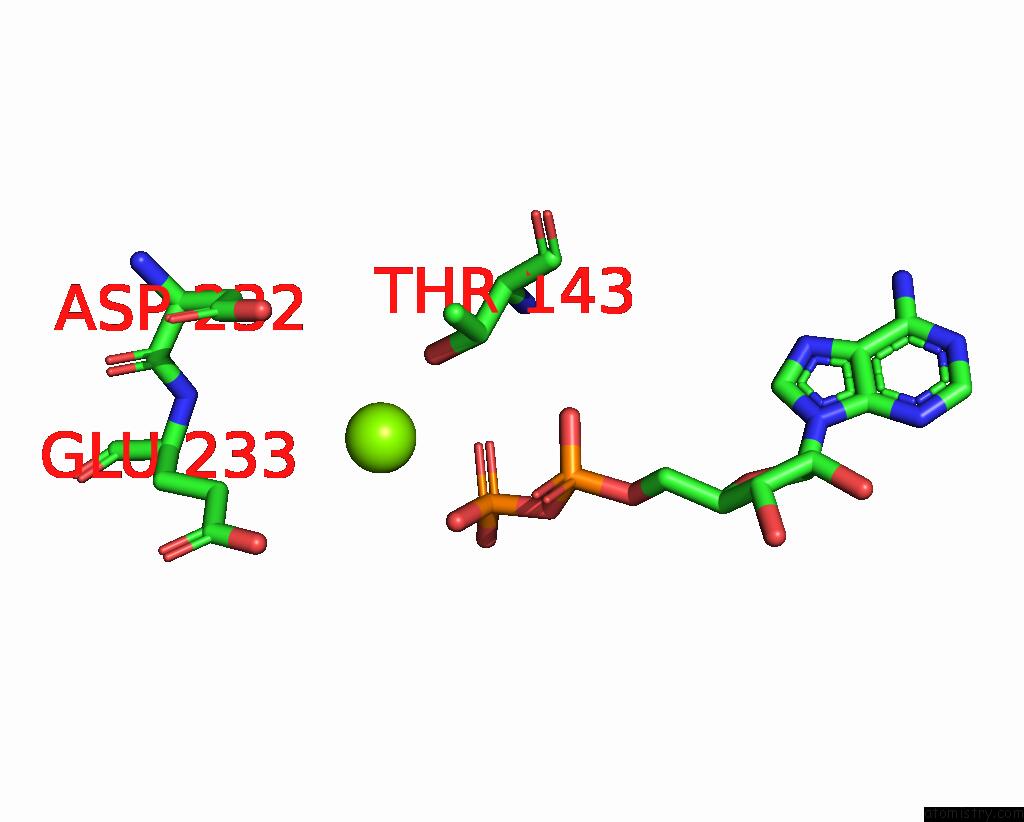
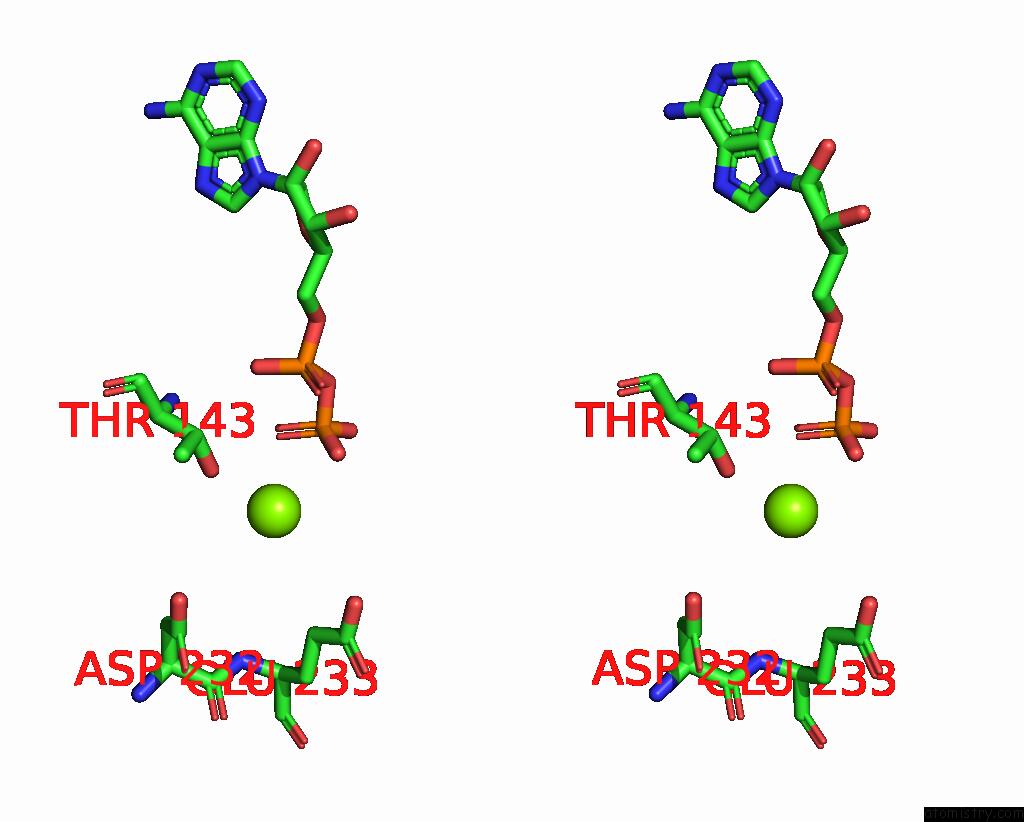
Magnesium binding site 3 out of 6 in 8vct
Go back to
Magnesium binding site 3 out
of 6 in the Cyoem Structure of the Tnsc(1-503)-Tnsd(1-318)-Dna Complex in A 6:2:1 Stoichiometry From E. Coli TN7 Bound to Atpgs and Adp
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 3 of Cyoem Structure of the Tnsc(1-503)-Tnsd(1-318)-Dna Complex in A 6:2:1 Stoichiometry From E. Coli TN7 Bound to Atpgs and Adp within 5.0Å range:
|
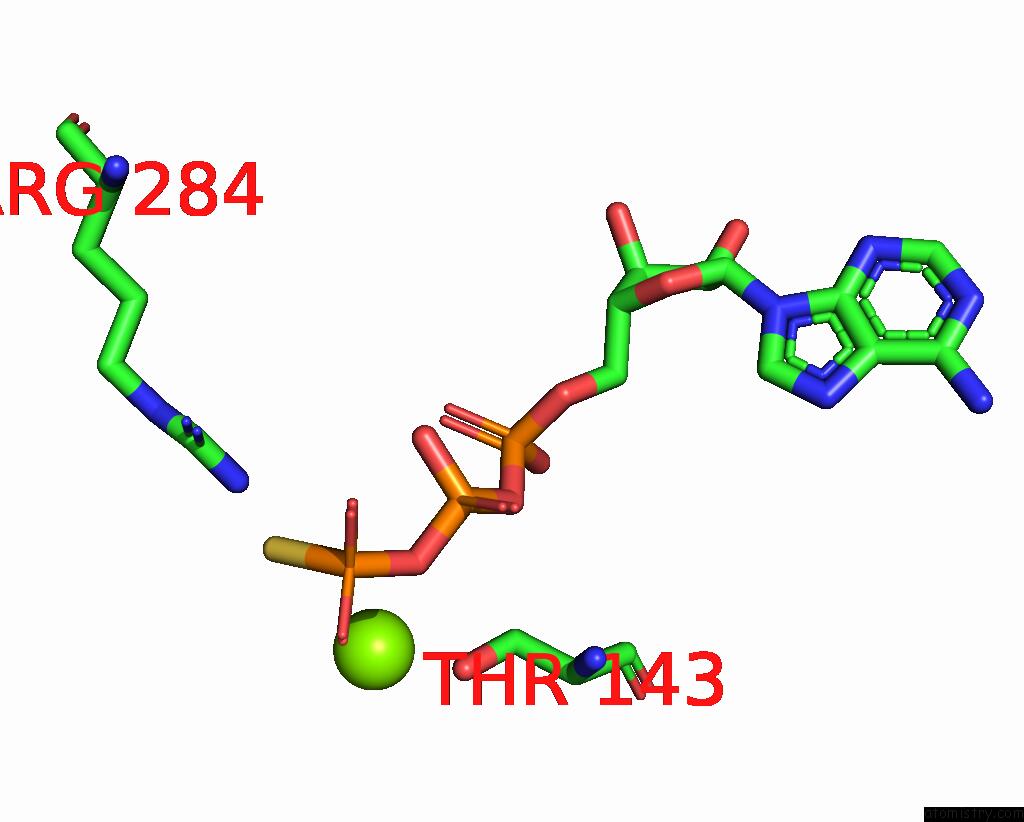
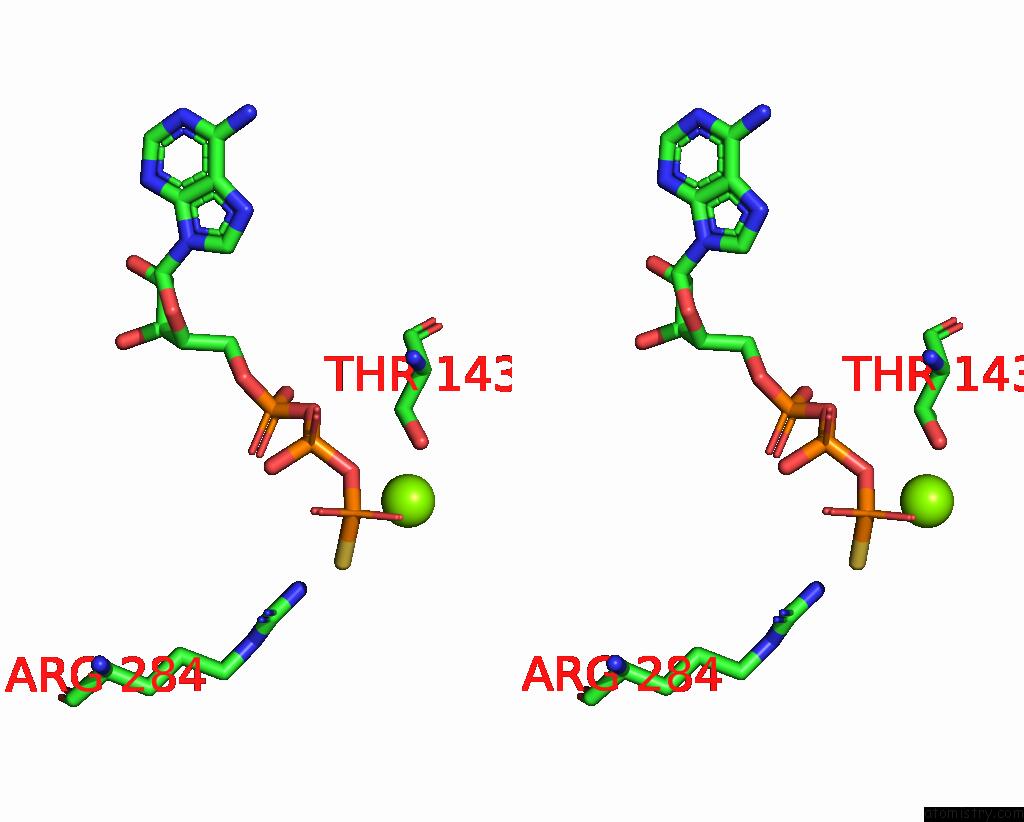
Magnesium binding site 4 out of 6 in 8vct
Go back to
Magnesium binding site 4 out
of 6 in the Cyoem Structure of the Tnsc(1-503)-Tnsd(1-318)-Dna Complex in A 6:2:1 Stoichiometry From E. Coli TN7 Bound to Atpgs and Adp
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 4 of Cyoem Structure of the Tnsc(1-503)-Tnsd(1-318)-Dna Complex in A 6:2:1 Stoichiometry From E. Coli TN7 Bound to Atpgs and Adp within 5.0Å range:
|
Magnesium binding site 5 out of 6 in 8vct
Go back to
Magnesium binding site 5 out
of 6 in the Cyoem Structure of the Tnsc(1-503)-Tnsd(1-318)-Dna Complex in A 6:2:1 Stoichiometry From E. Coli TN7 Bound to Atpgs and Adp
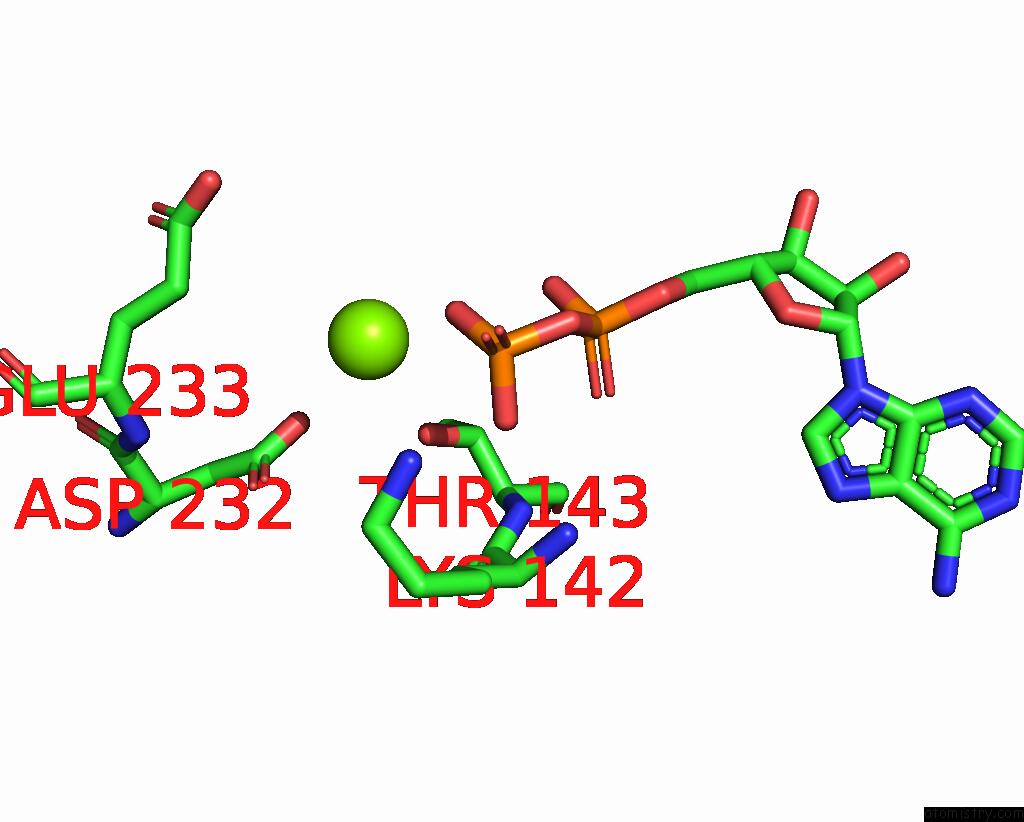
Mono view
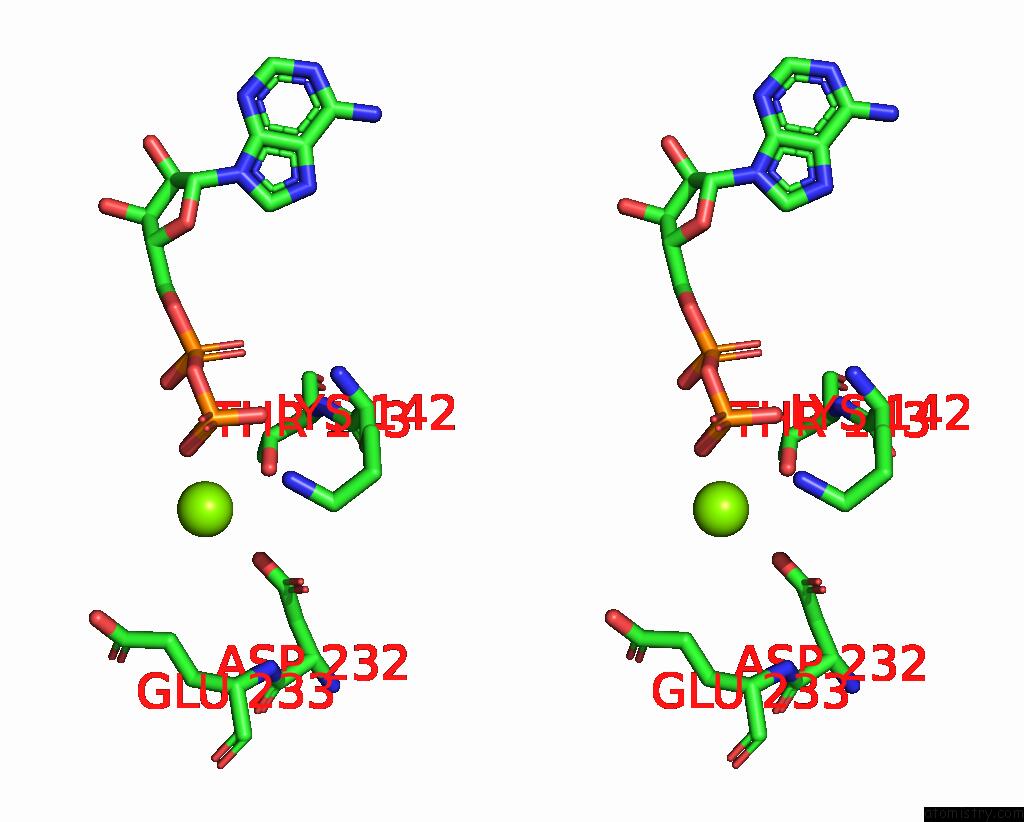
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 5 of Cyoem Structure of the Tnsc(1-503)-Tnsd(1-318)-Dna Complex in A 6:2:1 Stoichiometry From E. Coli TN7 Bound to Atpgs and Adp within 5.0Å range:
|
Magnesium binding site 6 out of 6 in 8vct
Go back to
Magnesium binding site 6 out
of 6 in the Cyoem Structure of the Tnsc(1-503)-Tnsd(1-318)-Dna Complex in A 6:2:1 Stoichiometry From E. Coli TN7 Bound to Atpgs and Adp
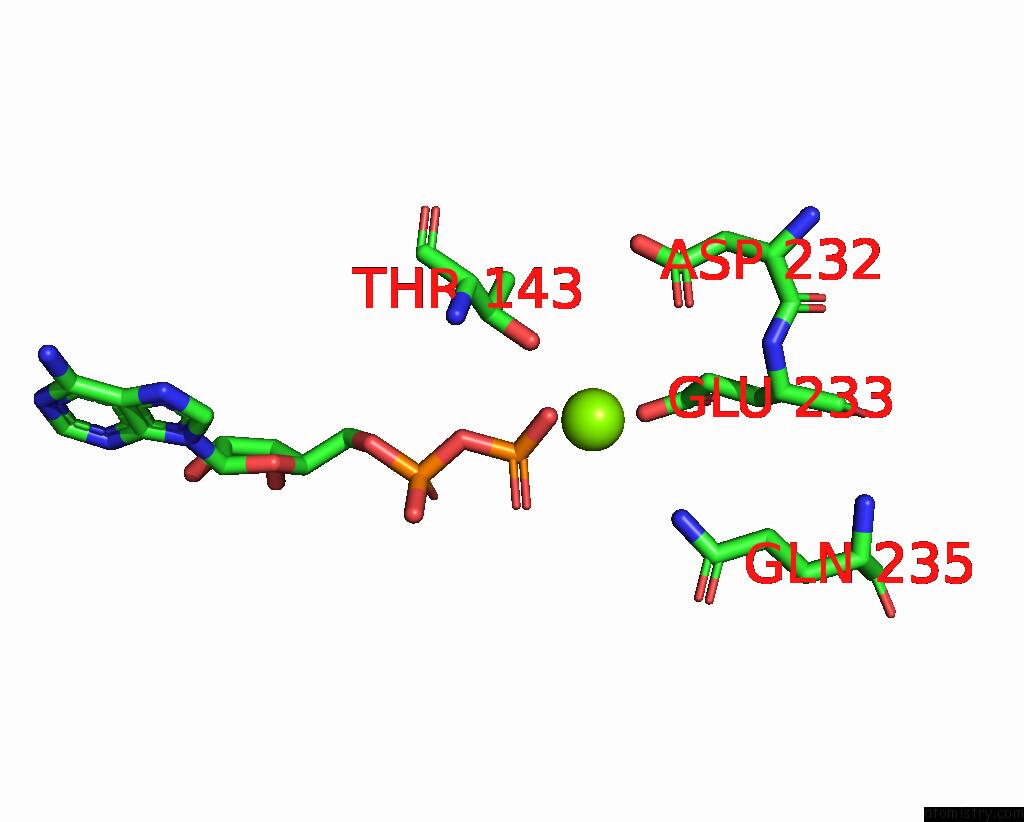
Mono view
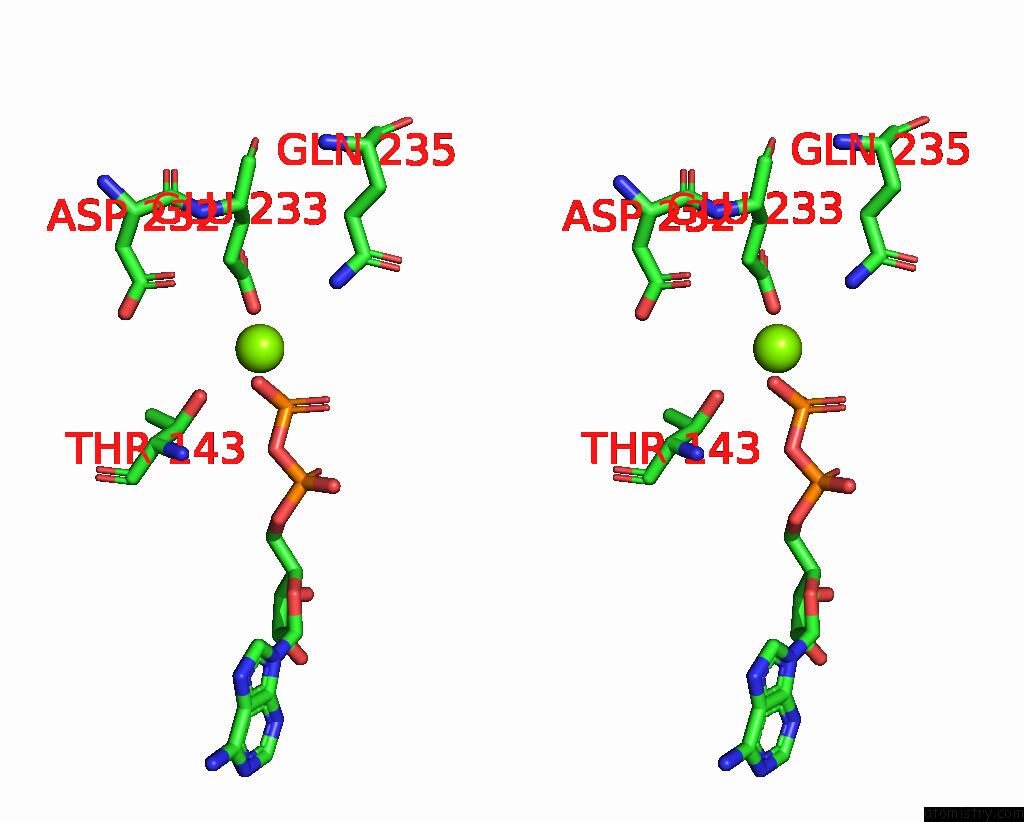
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 6 of Cyoem Structure of the Tnsc(1-503)-Tnsd(1-318)-Dna Complex in A 6:2:1 Stoichiometry From E. Coli TN7 Bound to Atpgs and Adp within 5.0Å range:
|
Reference:
Y.Shen,
S.S.Krishnan,
M.T.Petassi,
M.A.Hancock,
J.E.Peters,
A.Guarne.
Assembly of the TN7 Targeting Complex By A Regulated Stepwise Process. Mol.Cell V. 84 2368 2024.
ISSN: ISSN 1097-2765
PubMed: 38834067
DOI: 10.1016/J.MOLCEL.2024.05.012
Page generated: Fri Aug 15 17:35:41 2025
ISSN: ISSN 1097-2765
PubMed: 38834067
DOI: 10.1016/J.MOLCEL.2024.05.012
Last articles
Mg in 9F37Mg in 9EZL
Mg in 9F28
Mg in 9F2R
Mg in 9F12
Mg in 9F11
Mg in 9F10
Mg in 9F0Z
Mg in 9F0Y
Mg in 9F07