Magnesium »
PDB 8vbi-8vn3 »
8vgb »
Magnesium in PDB 8vgb: Crystal Structure of Guanine Nucleotide-Binding Protein Alpha Subunit (G Protein) From Oryza Sativa in Complex with Gdp
Protein crystallography data
The structure of Crystal Structure of Guanine Nucleotide-Binding Protein Alpha Subunit (G Protein) From Oryza Sativa in Complex with Gdp, PDB code: 8vgb
was solved by
S.G.Lee,
J.M.Jez,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 48.37 / 2.99 |
| Space group | P 1 21 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 68.393, 68.419, 167.977, 90, 89.91, 90 |
| R / Rfree (%) | 26.6 / 30.6 |
Magnesium Binding Sites:
The binding sites of Magnesium atom in the Crystal Structure of Guanine Nucleotide-Binding Protein Alpha Subunit (G Protein) From Oryza Sativa in Complex with Gdp
(pdb code 8vgb). This binding sites where shown within
5.0 Angstroms radius around Magnesium atom.
In total 4 binding sites of Magnesium where determined in the Crystal Structure of Guanine Nucleotide-Binding Protein Alpha Subunit (G Protein) From Oryza Sativa in Complex with Gdp, PDB code: 8vgb:
Jump to Magnesium binding site number: 1; 2; 3; 4;
In total 4 binding sites of Magnesium where determined in the Crystal Structure of Guanine Nucleotide-Binding Protein Alpha Subunit (G Protein) From Oryza Sativa in Complex with Gdp, PDB code: 8vgb:
Jump to Magnesium binding site number: 1; 2; 3; 4;
Magnesium binding site 1 out of 4 in 8vgb
Go back to
Magnesium binding site 1 out
of 4 in the Crystal Structure of Guanine Nucleotide-Binding Protein Alpha Subunit (G Protein) From Oryza Sativa in Complex with Gdp
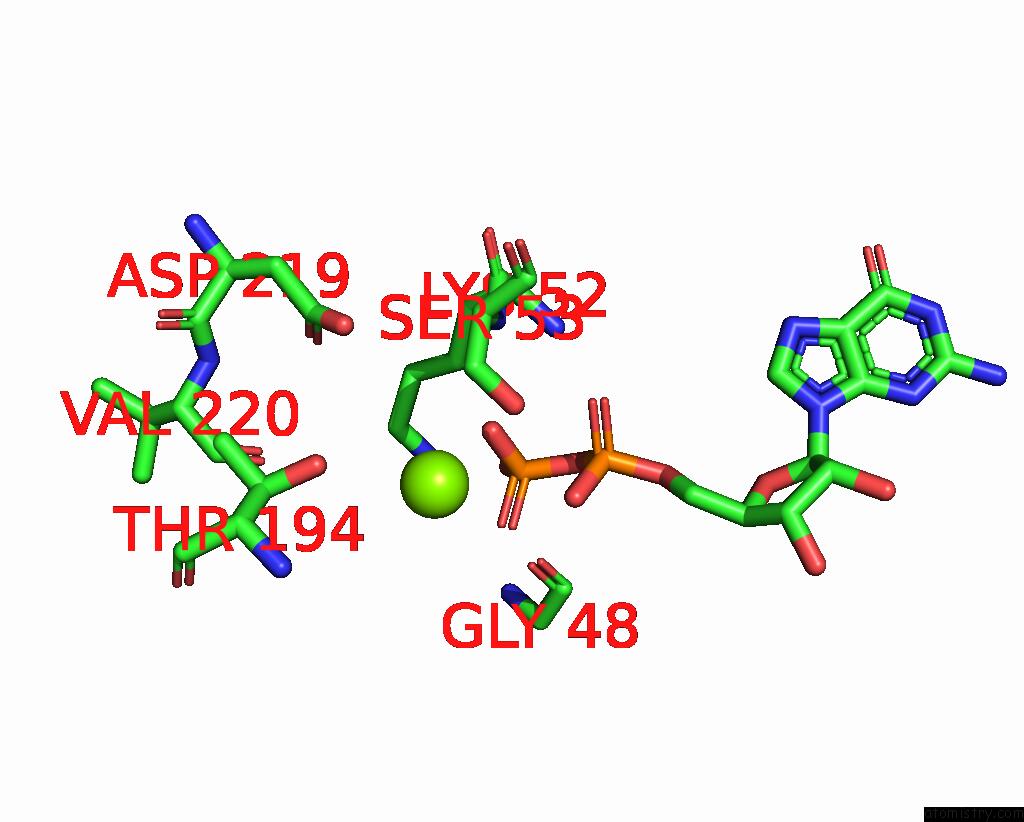
Mono view
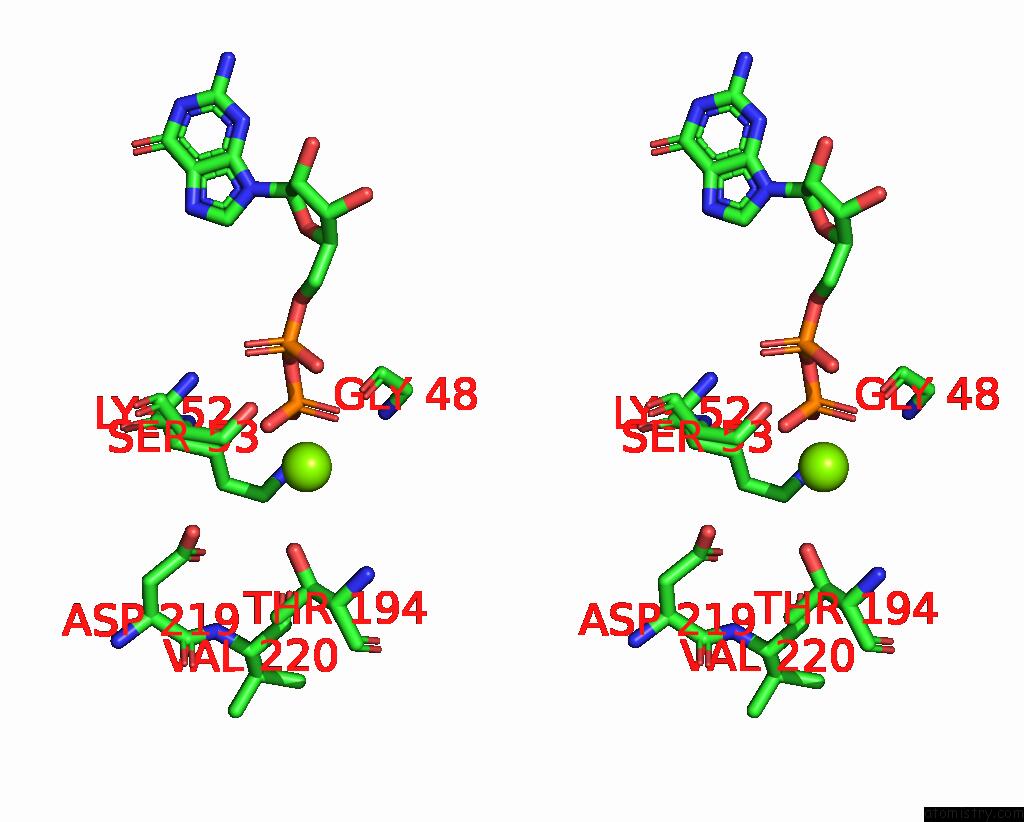
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 1 of Crystal Structure of Guanine Nucleotide-Binding Protein Alpha Subunit (G Protein) From Oryza Sativa in Complex with Gdp within 5.0Å range:
|
Magnesium binding site 2 out of 4 in 8vgb
Go back to
Magnesium binding site 2 out
of 4 in the Crystal Structure of Guanine Nucleotide-Binding Protein Alpha Subunit (G Protein) From Oryza Sativa in Complex with Gdp
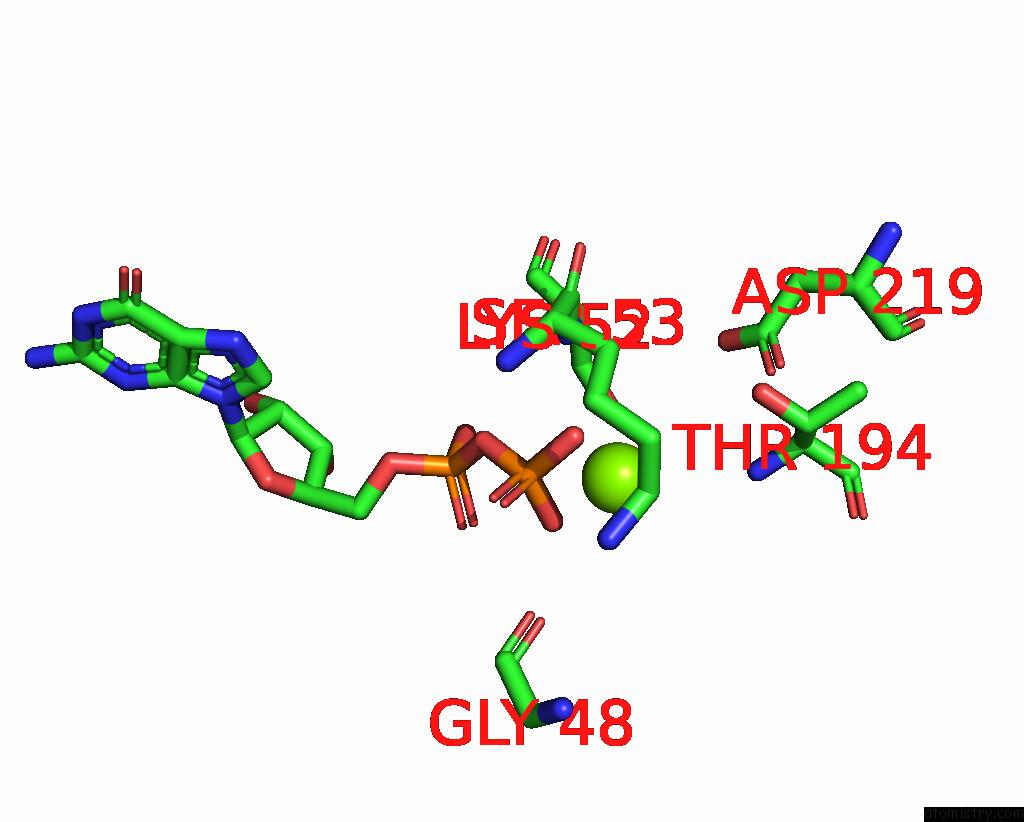
Mono view
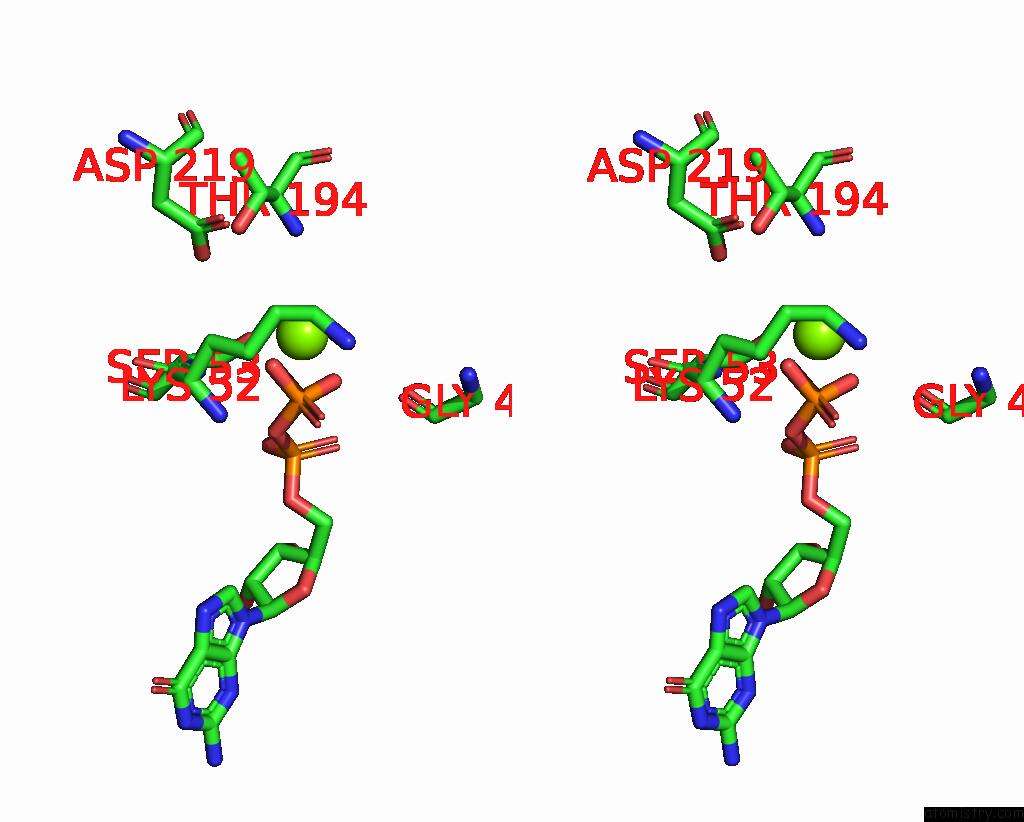
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 2 of Crystal Structure of Guanine Nucleotide-Binding Protein Alpha Subunit (G Protein) From Oryza Sativa in Complex with Gdp within 5.0Å range:
|
Magnesium binding site 3 out of 4 in 8vgb
Go back to
Magnesium binding site 3 out
of 4 in the Crystal Structure of Guanine Nucleotide-Binding Protein Alpha Subunit (G Protein) From Oryza Sativa in Complex with Gdp
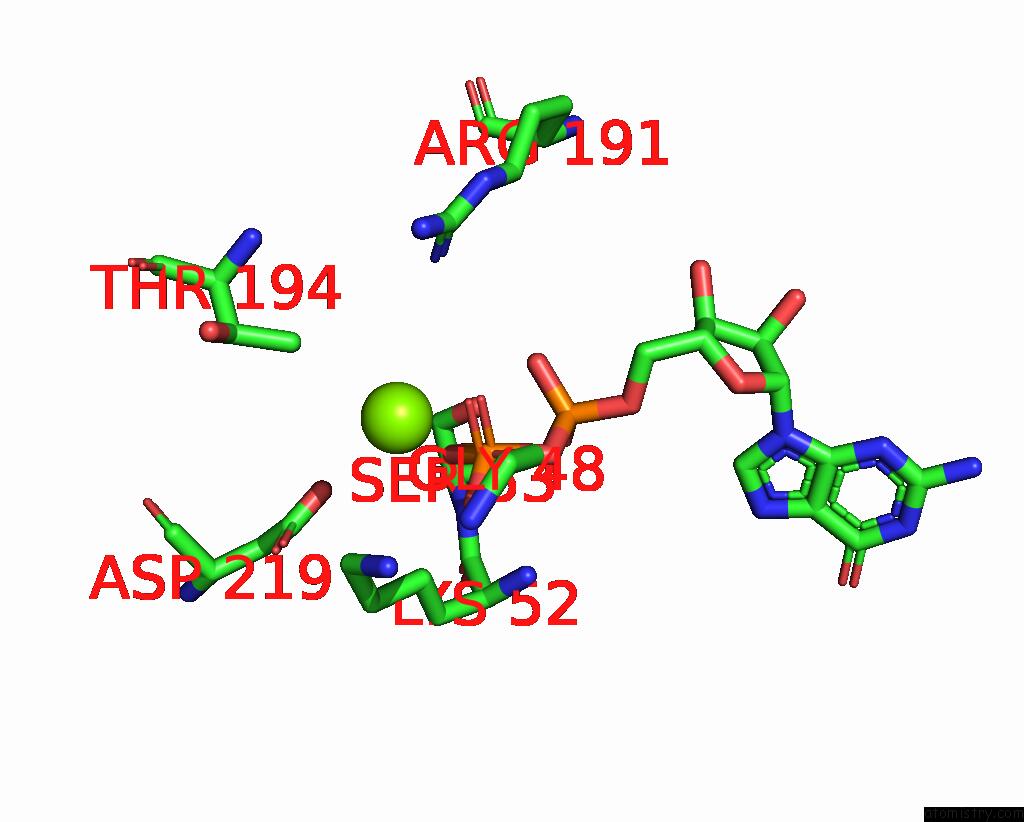
Mono view
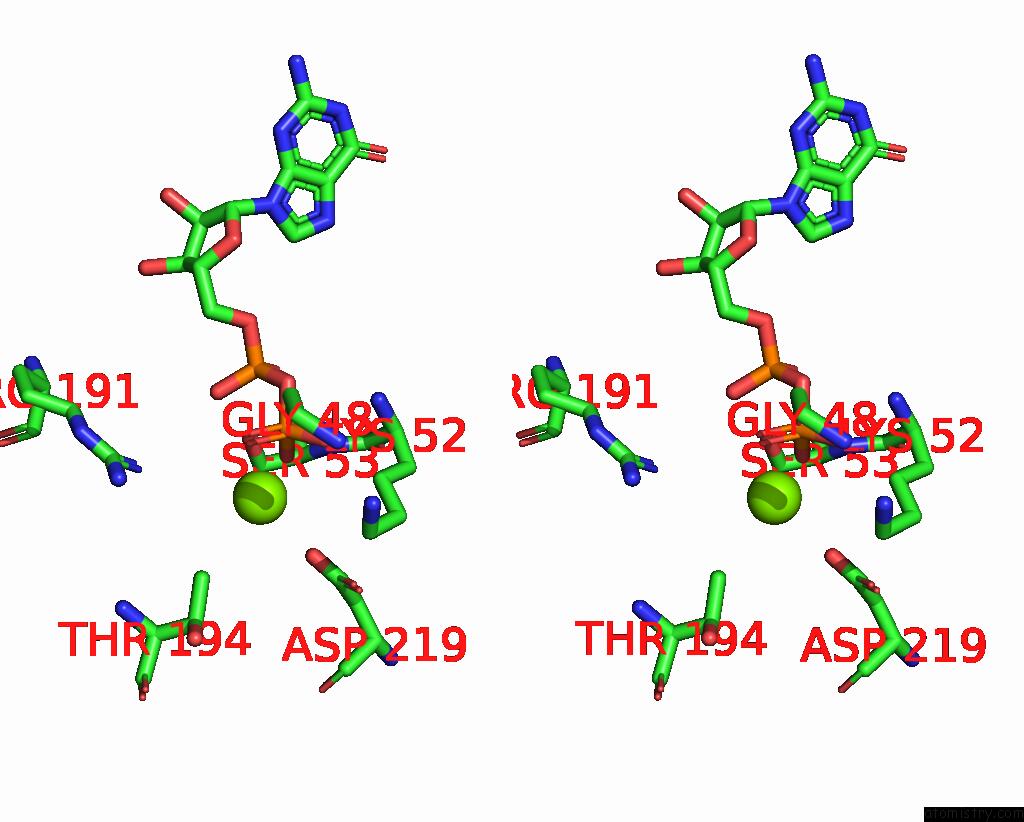
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 3 of Crystal Structure of Guanine Nucleotide-Binding Protein Alpha Subunit (G Protein) From Oryza Sativa in Complex with Gdp within 5.0Å range:
|
Magnesium binding site 4 out of 4 in 8vgb
Go back to
Magnesium binding site 4 out
of 4 in the Crystal Structure of Guanine Nucleotide-Binding Protein Alpha Subunit (G Protein) From Oryza Sativa in Complex with Gdp
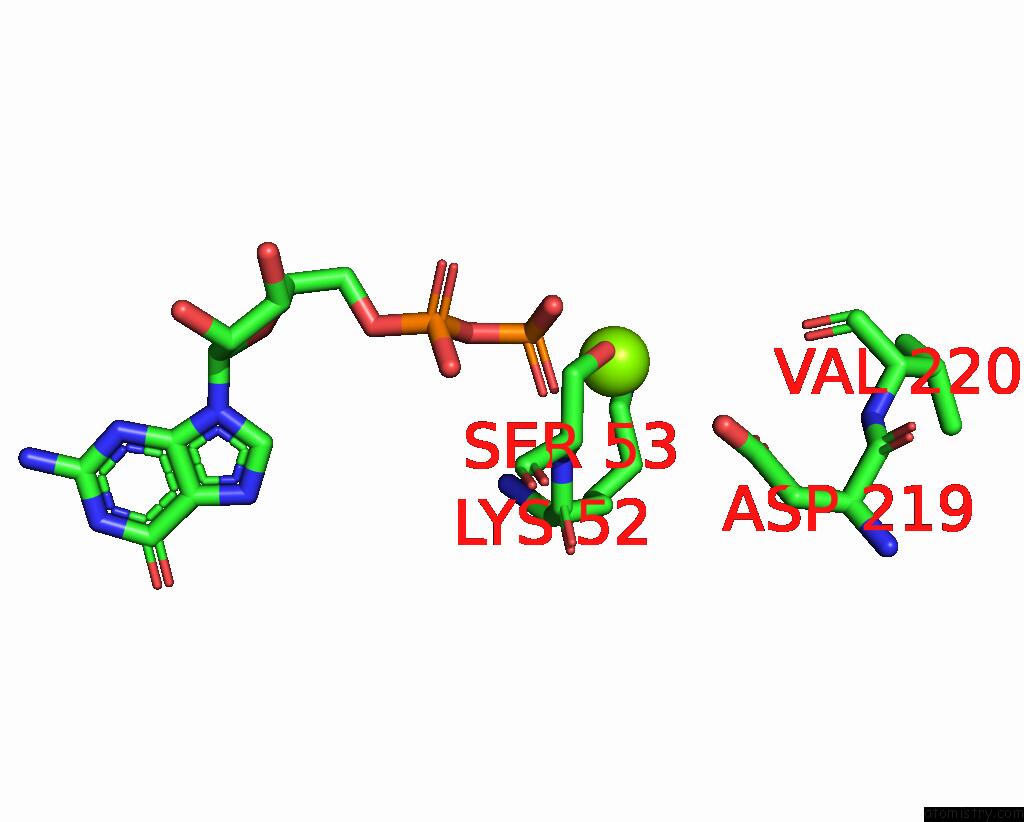
Mono view
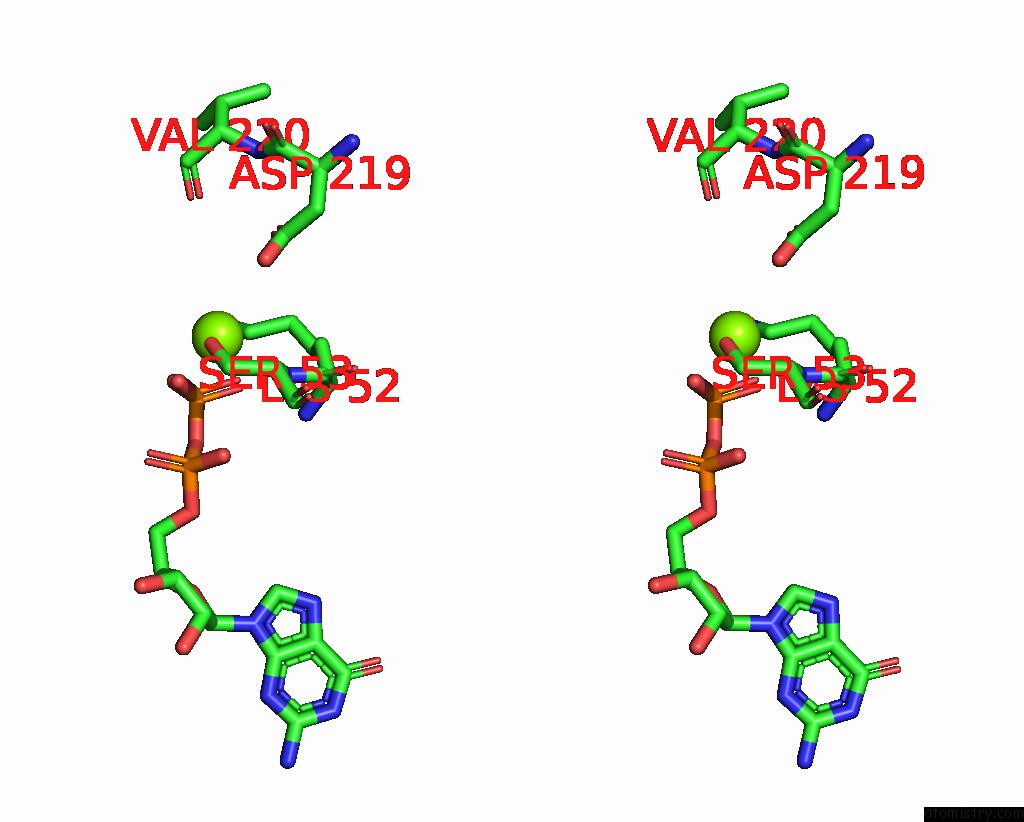
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 4 of Crystal Structure of Guanine Nucleotide-Binding Protein Alpha Subunit (G Protein) From Oryza Sativa in Complex with Gdp within 5.0Å range:
|
Reference:
M.D.Torres-Rodriguez,
S.G.Lee,
S.Roy Choudhury,
R.Paul,
B.Selvam,
D.Shukla,
J.M.Jez,
S.Pandey.
Structure-Function Analysis of Plant G-Protein Regulatory Mechanisms Identifies Key G Alpha-Rgs Protein Interactions. J.Biol.Chem. V. 300 07252 2024.
ISSN: ESSN 1083-351X
PubMed: 38569936
DOI: 10.1016/J.JBC.2024.107252
Page generated: Fri Aug 15 17:36:53 2025
ISSN: ESSN 1083-351X
PubMed: 38569936
DOI: 10.1016/J.JBC.2024.107252
Last articles
Mg in 8Z2LMg in 8YZX
Mg in 8YZU
Mg in 8Z1R
Mg in 8YZY
Mg in 8Z0J
Mg in 8Z0I
Mg in 8Z03
Mg in 8YWQ
Mg in 8YWI