Magnesium »
PDB 8xw7-8y9l »
8y7s »
Magnesium in PDB 8y7s: Crystal Structure of A Benzaldehyde Lyase Mutant M6 From Herbiconiux Sp. Salv-R1
Protein crystallography data
The structure of Crystal Structure of A Benzaldehyde Lyase Mutant M6 From Herbiconiux Sp. Salv-R1, PDB code: 8y7s
was solved by
Y.Li,
Y.F.Zhang,
Y.Y.Chen,
W.D.Liu,
P.Y.Yao,
Q.Q.Wu,
D.M.Zhu,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 29.19 / 2.68 |
| Space group | C 1 2 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 333.789, 98.696, 231.654, 90, 108.05, 90 |
| R / Rfree (%) | 18.9 / 23.6 |
Magnesium Binding Sites:
Pages:
>>> Page 1 <<< Page 2, Binding sites: 11 - 12;Binding sites:
The binding sites of Magnesium atom in the Crystal Structure of A Benzaldehyde Lyase Mutant M6 From Herbiconiux Sp. Salv-R1 (pdb code 8y7s). This binding sites where shown within 5.0 Angstroms radius around Magnesium atom.In total 12 binding sites of Magnesium where determined in the Crystal Structure of A Benzaldehyde Lyase Mutant M6 From Herbiconiux Sp. Salv-R1, PDB code: 8y7s:
Jump to Magnesium binding site number: 1; 2; 3; 4; 5; 6; 7; 8; 9; 10;
Magnesium binding site 1 out of 12 in 8y7s
Go back to
Magnesium binding site 1 out
of 12 in the Crystal Structure of A Benzaldehyde Lyase Mutant M6 From Herbiconiux Sp. Salv-R1
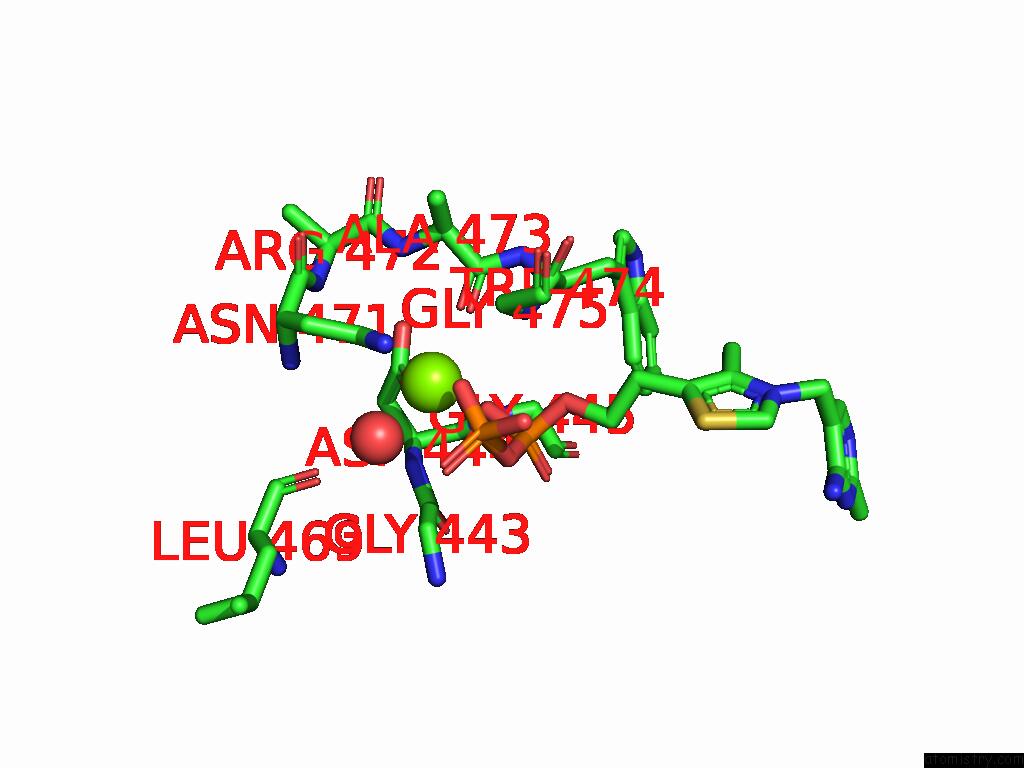
Mono view
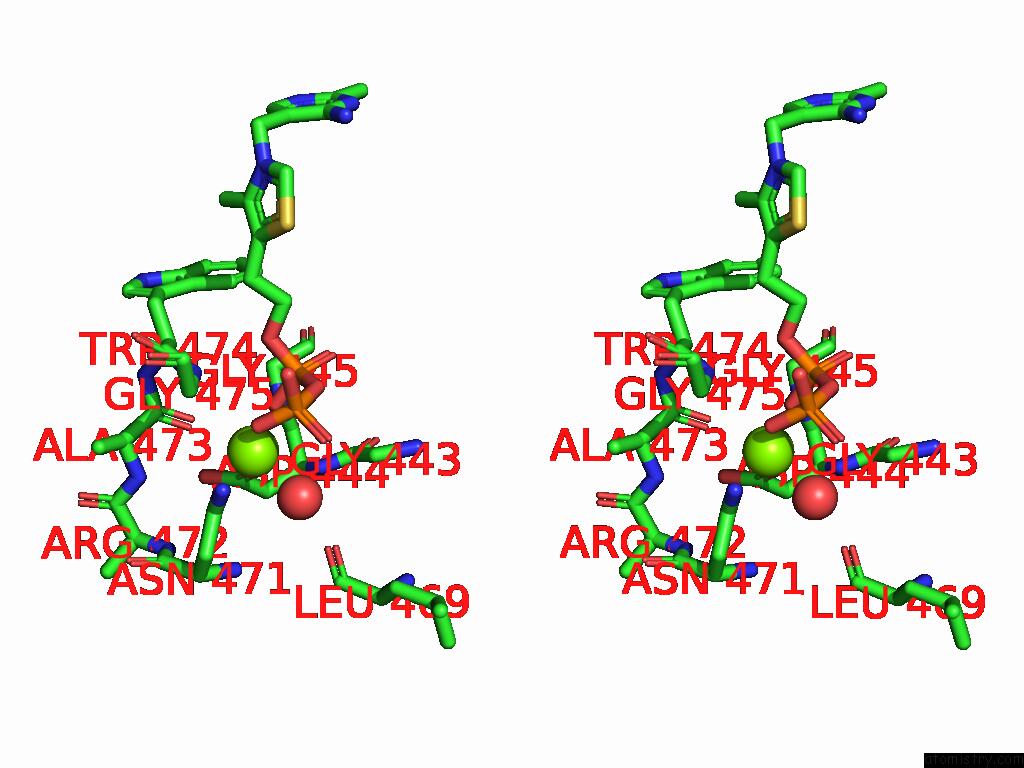
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 1 of Crystal Structure of A Benzaldehyde Lyase Mutant M6 From Herbiconiux Sp. Salv-R1 within 5.0Å range:
|
Magnesium binding site 2 out of 12 in 8y7s
Go back to
Magnesium binding site 2 out
of 12 in the Crystal Structure of A Benzaldehyde Lyase Mutant M6 From Herbiconiux Sp. Salv-R1
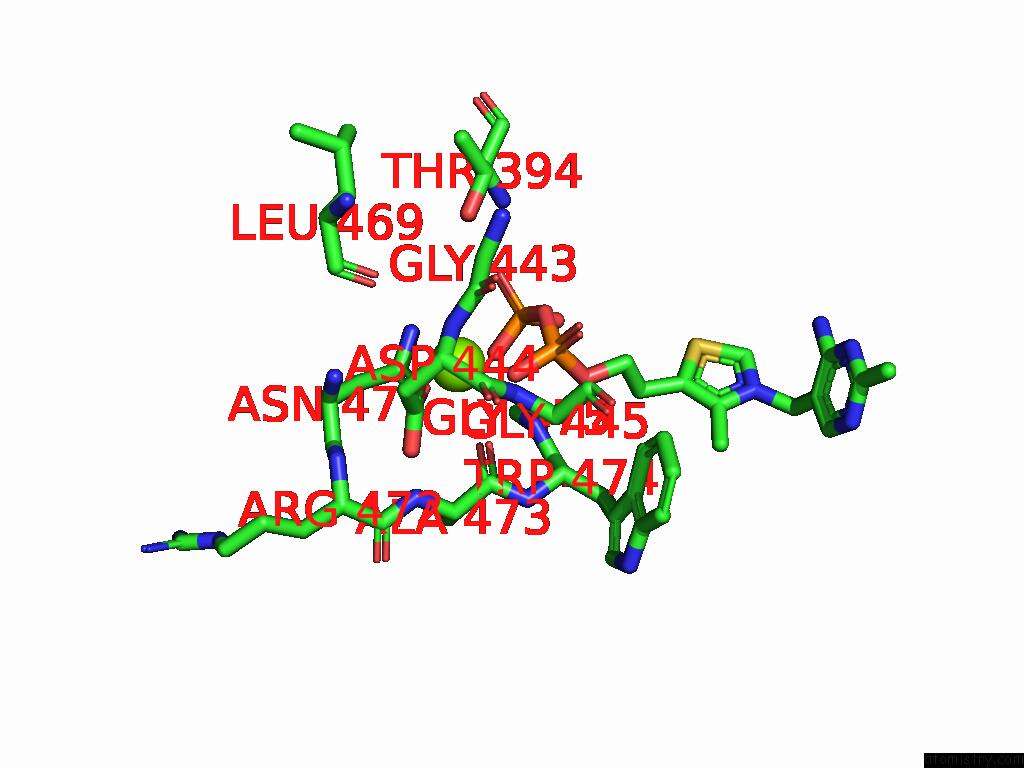
Mono view
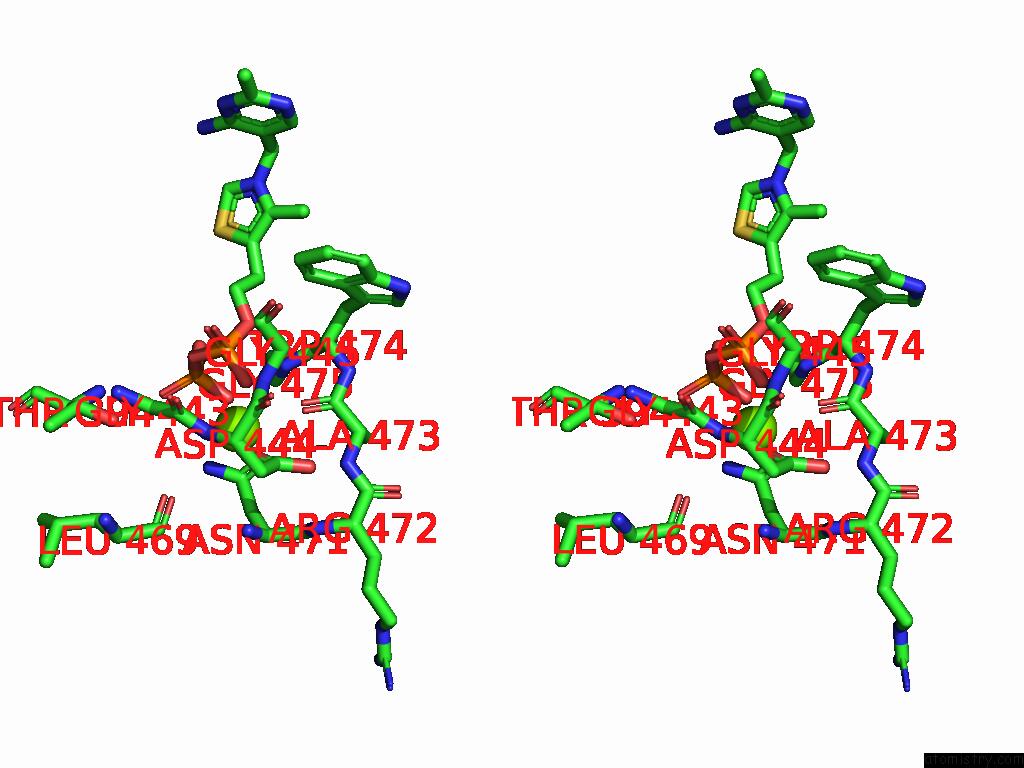
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 2 of Crystal Structure of A Benzaldehyde Lyase Mutant M6 From Herbiconiux Sp. Salv-R1 within 5.0Å range:
|
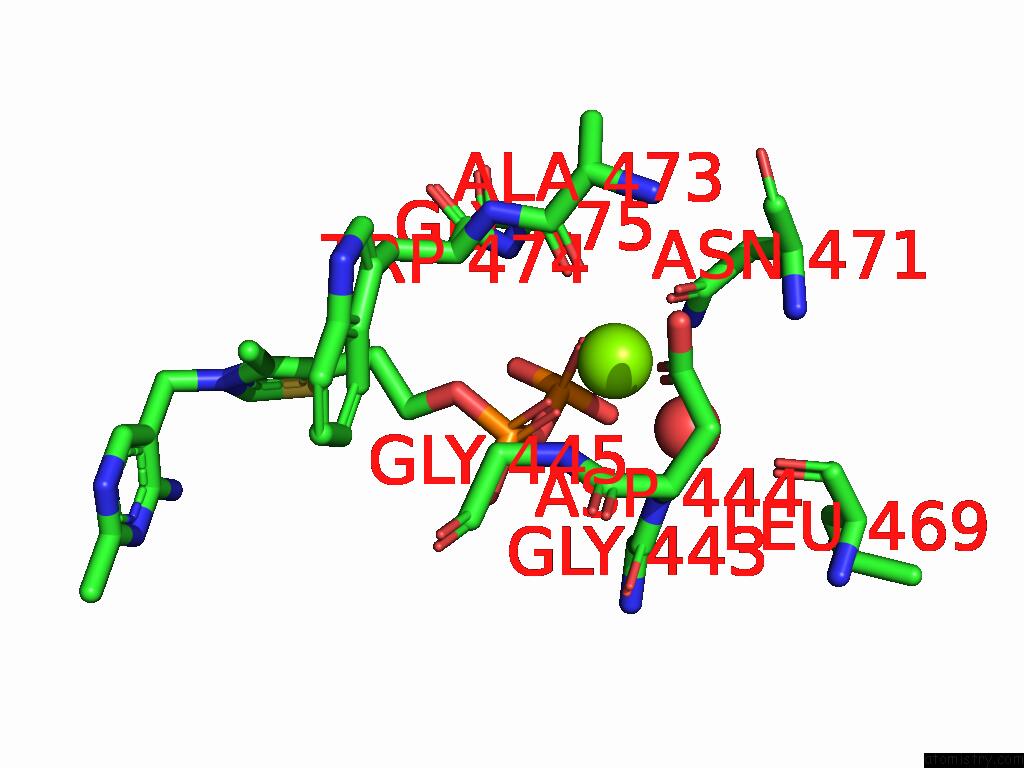
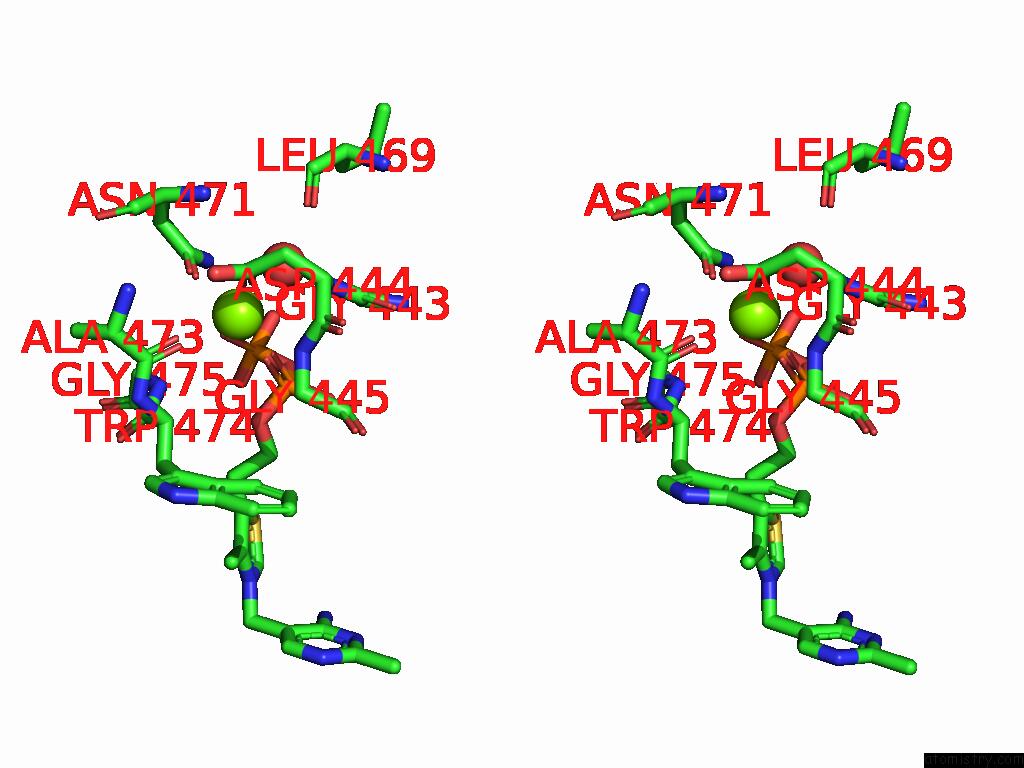
Magnesium binding site 3 out of 12 in 8y7s
Go back to
Magnesium binding site 3 out
of 12 in the Crystal Structure of A Benzaldehyde Lyase Mutant M6 From Herbiconiux Sp. Salv-R1
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 3 of Crystal Structure of A Benzaldehyde Lyase Mutant M6 From Herbiconiux Sp. Salv-R1 within 5.0Å range:
|
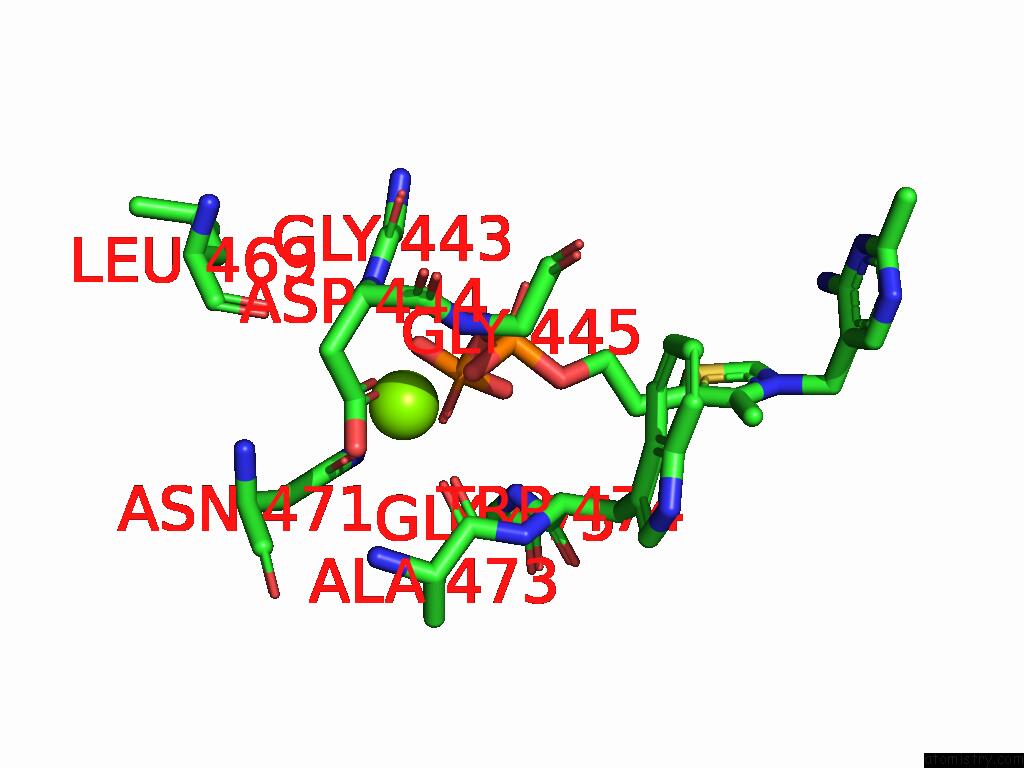
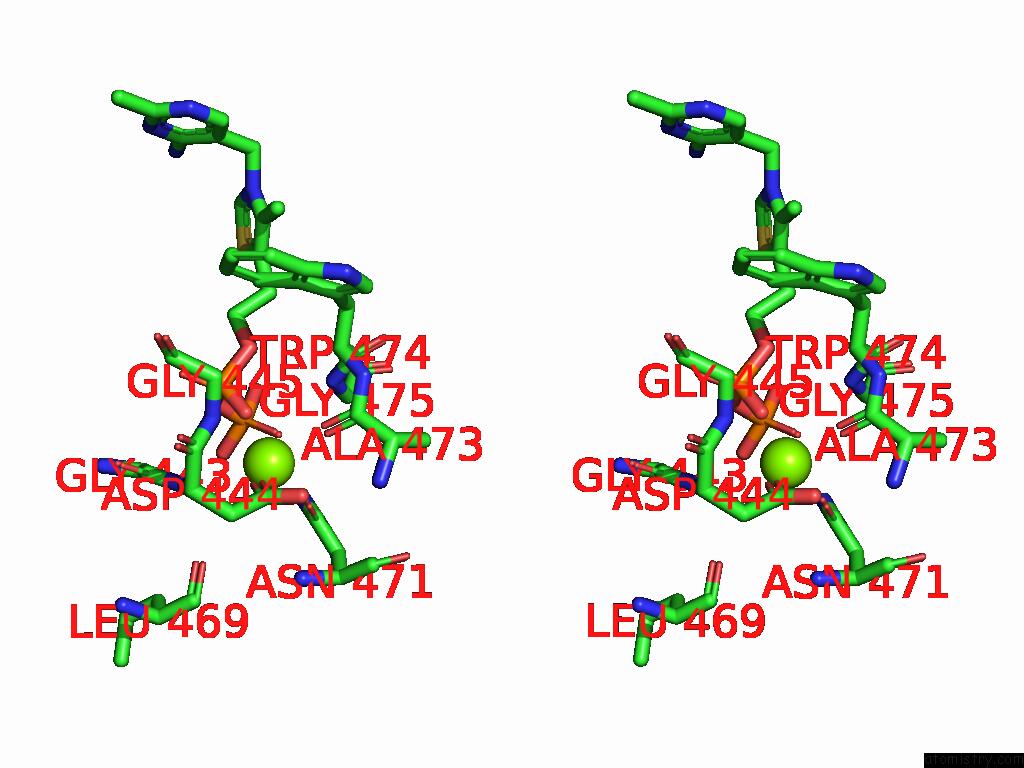
Magnesium binding site 4 out of 12 in 8y7s
Go back to
Magnesium binding site 4 out
of 12 in the Crystal Structure of A Benzaldehyde Lyase Mutant M6 From Herbiconiux Sp. Salv-R1
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 4 of Crystal Structure of A Benzaldehyde Lyase Mutant M6 From Herbiconiux Sp. Salv-R1 within 5.0Å range:
|
Magnesium binding site 5 out of 12 in 8y7s
Go back to
Magnesium binding site 5 out
of 12 in the Crystal Structure of A Benzaldehyde Lyase Mutant M6 From Herbiconiux Sp. Salv-R1
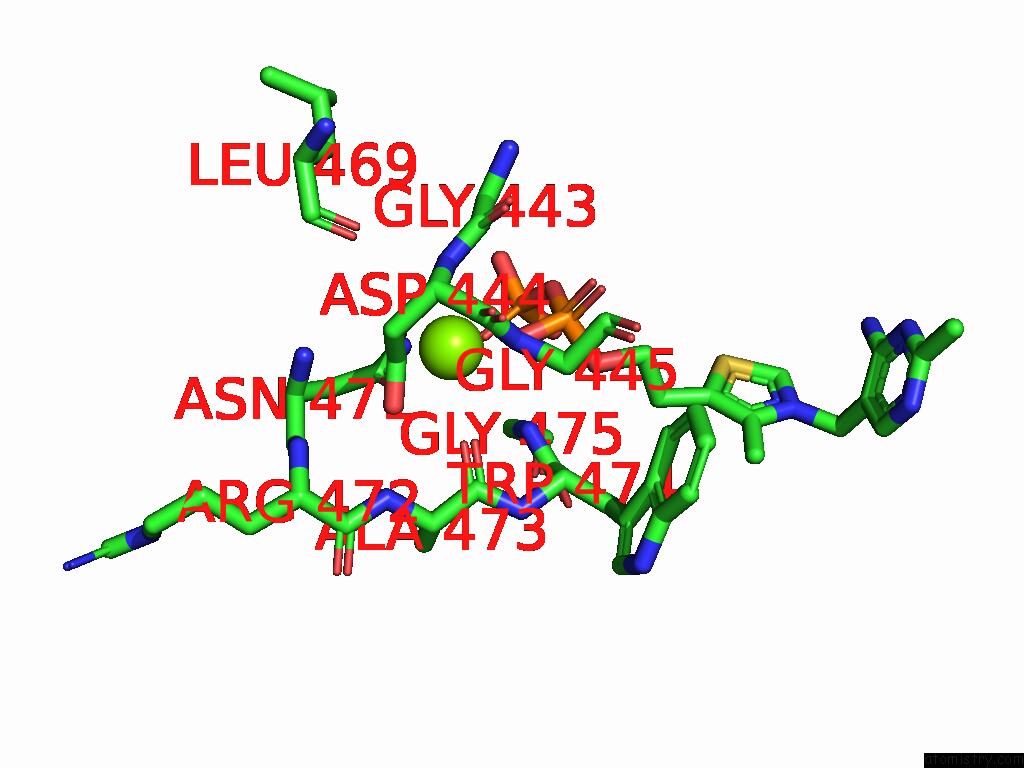
Mono view
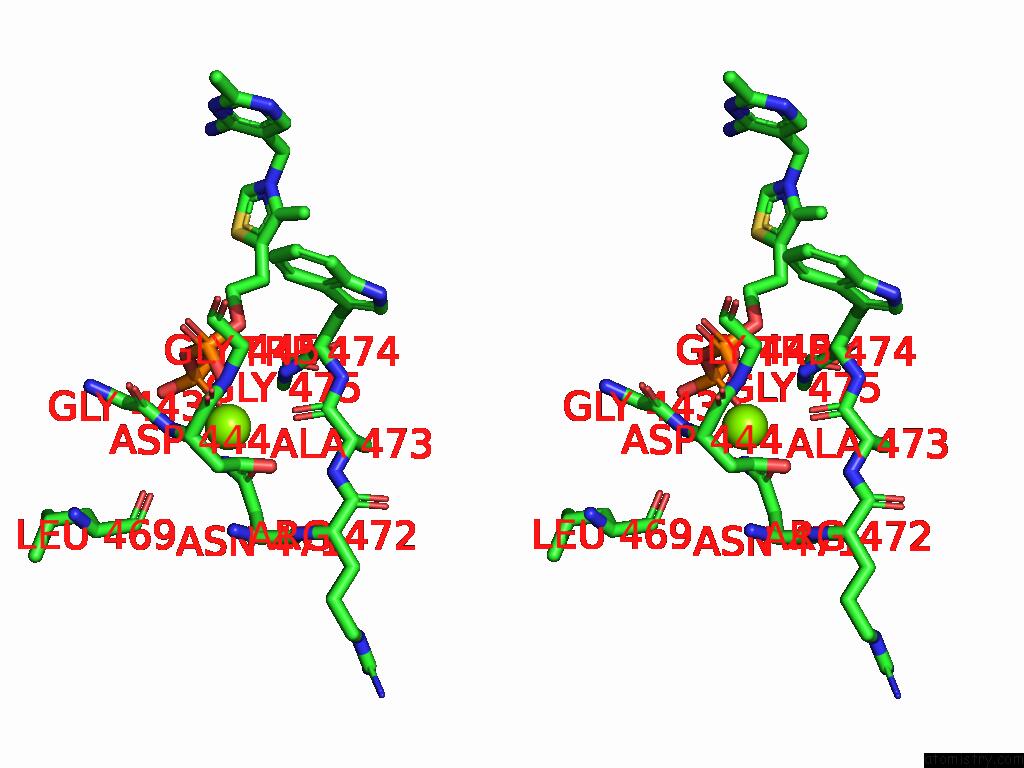
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 5 of Crystal Structure of A Benzaldehyde Lyase Mutant M6 From Herbiconiux Sp. Salv-R1 within 5.0Å range:
|
Magnesium binding site 6 out of 12 in 8y7s
Go back to
Magnesium binding site 6 out
of 12 in the Crystal Structure of A Benzaldehyde Lyase Mutant M6 From Herbiconiux Sp. Salv-R1

Mono view
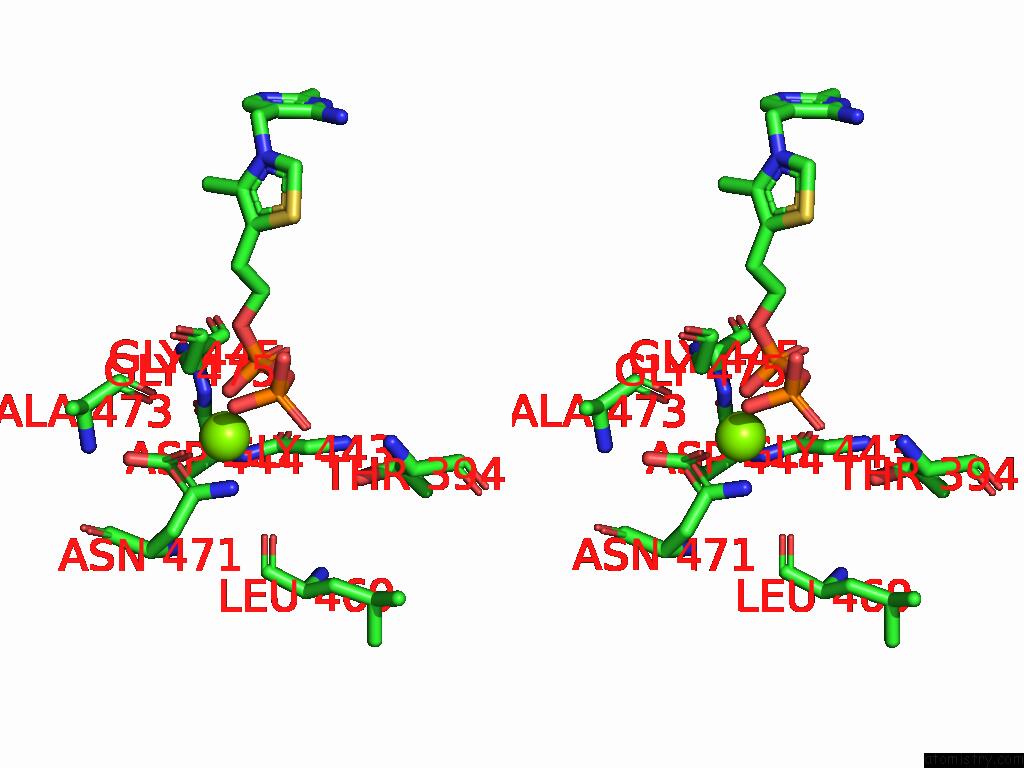
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 6 of Crystal Structure of A Benzaldehyde Lyase Mutant M6 From Herbiconiux Sp. Salv-R1 within 5.0Å range:
|
Magnesium binding site 7 out of 12 in 8y7s
Go back to
Magnesium binding site 7 out
of 12 in the Crystal Structure of A Benzaldehyde Lyase Mutant M6 From Herbiconiux Sp. Salv-R1
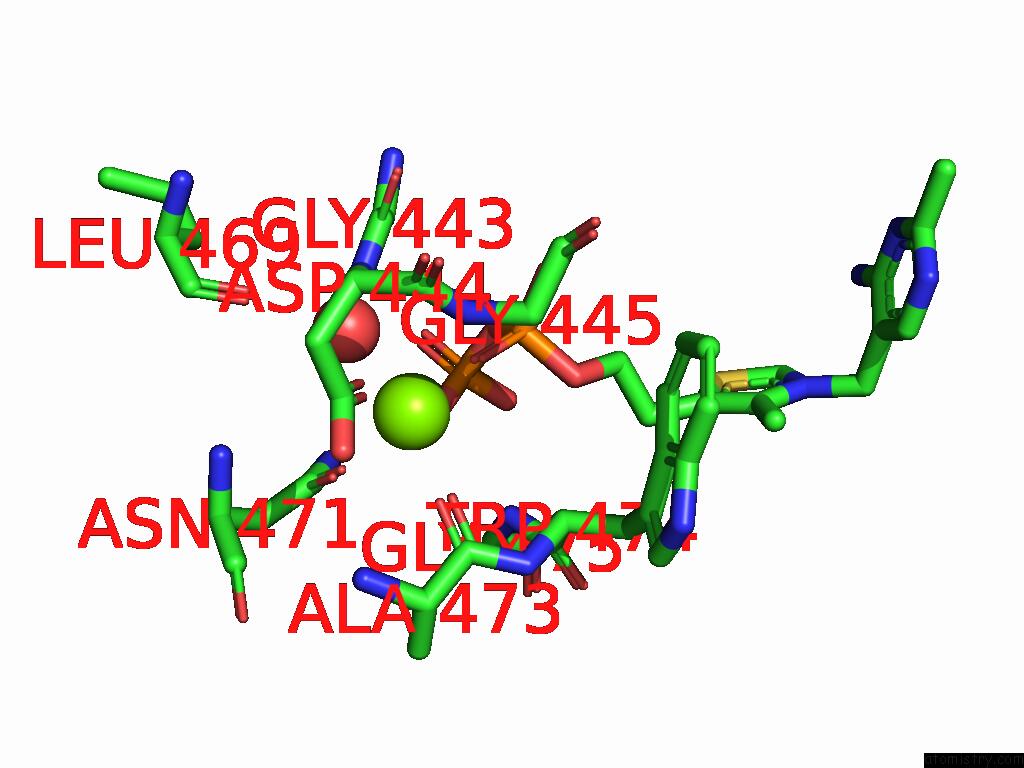
Mono view
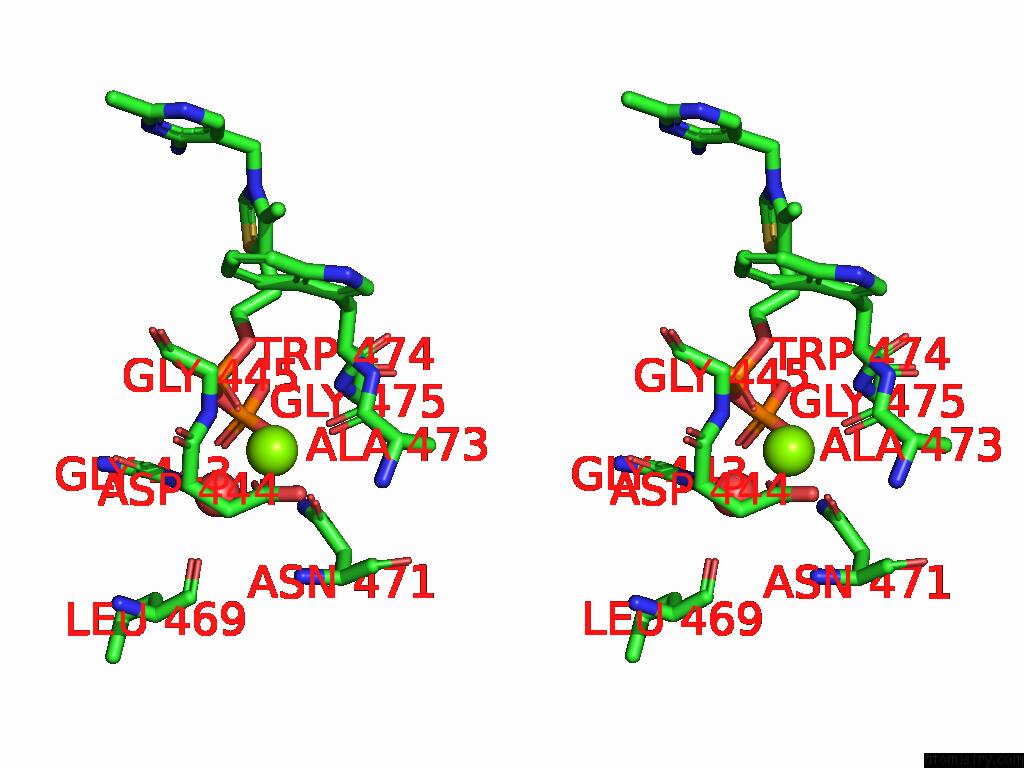
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 7 of Crystal Structure of A Benzaldehyde Lyase Mutant M6 From Herbiconiux Sp. Salv-R1 within 5.0Å range:
|
Magnesium binding site 8 out of 12 in 8y7s
Go back to
Magnesium binding site 8 out
of 12 in the Crystal Structure of A Benzaldehyde Lyase Mutant M6 From Herbiconiux Sp. Salv-R1
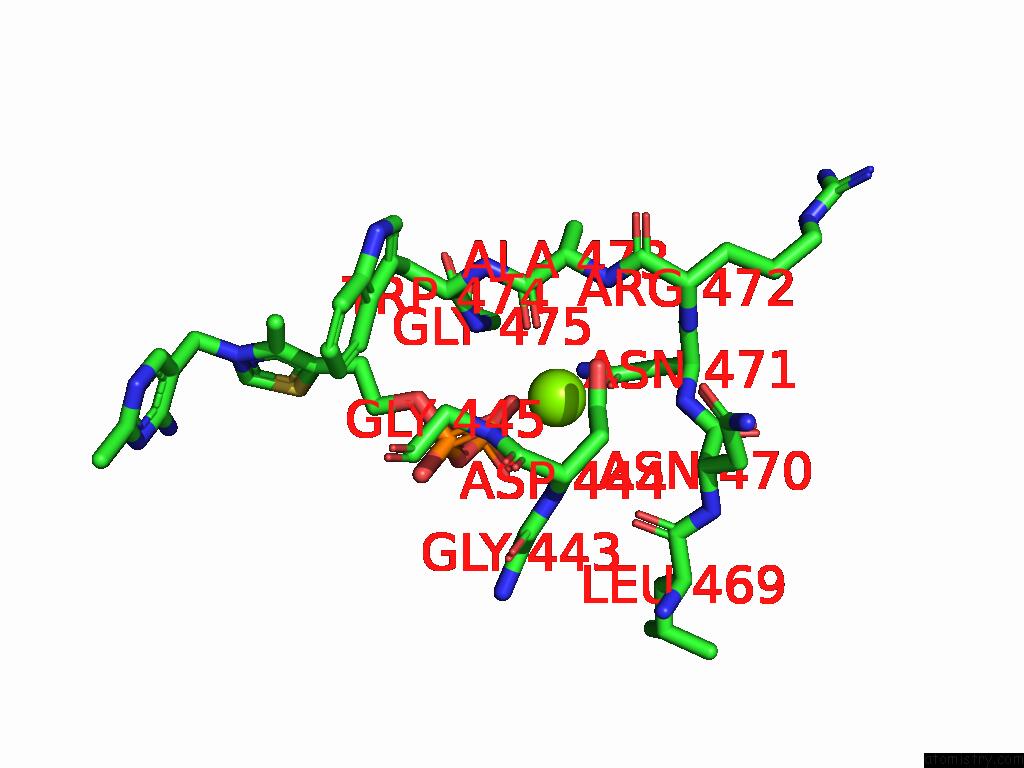
Mono view
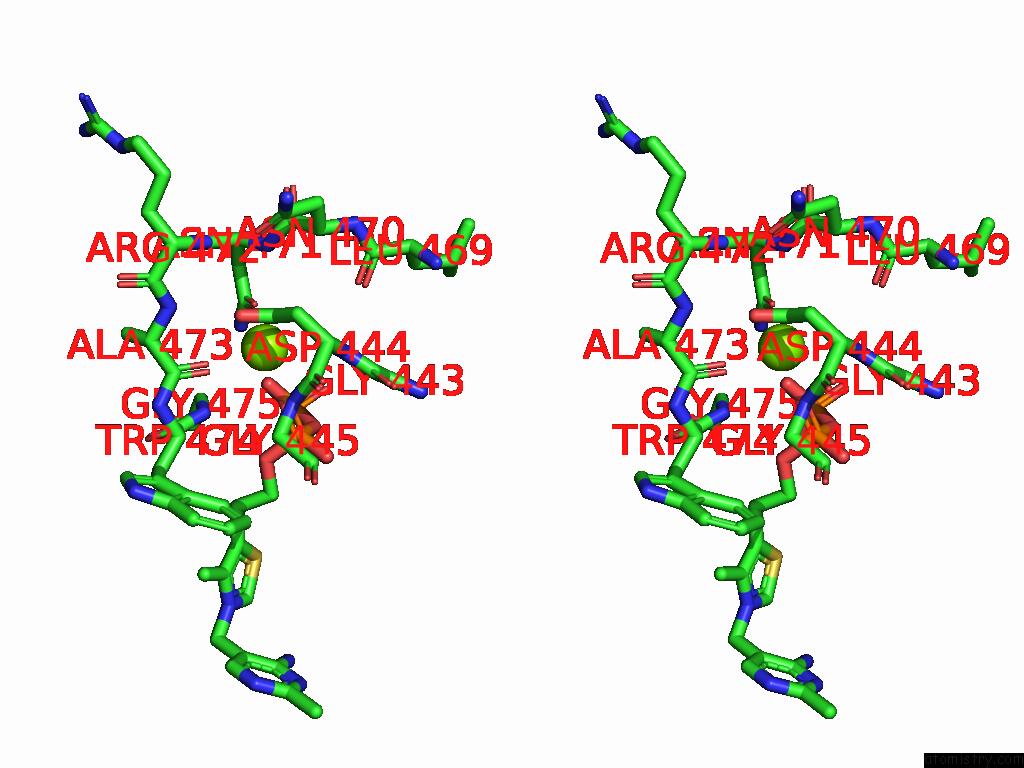
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 8 of Crystal Structure of A Benzaldehyde Lyase Mutant M6 From Herbiconiux Sp. Salv-R1 within 5.0Å range:
|
Magnesium binding site 9 out of 12 in 8y7s
Go back to
Magnesium binding site 9 out
of 12 in the Crystal Structure of A Benzaldehyde Lyase Mutant M6 From Herbiconiux Sp. Salv-R1
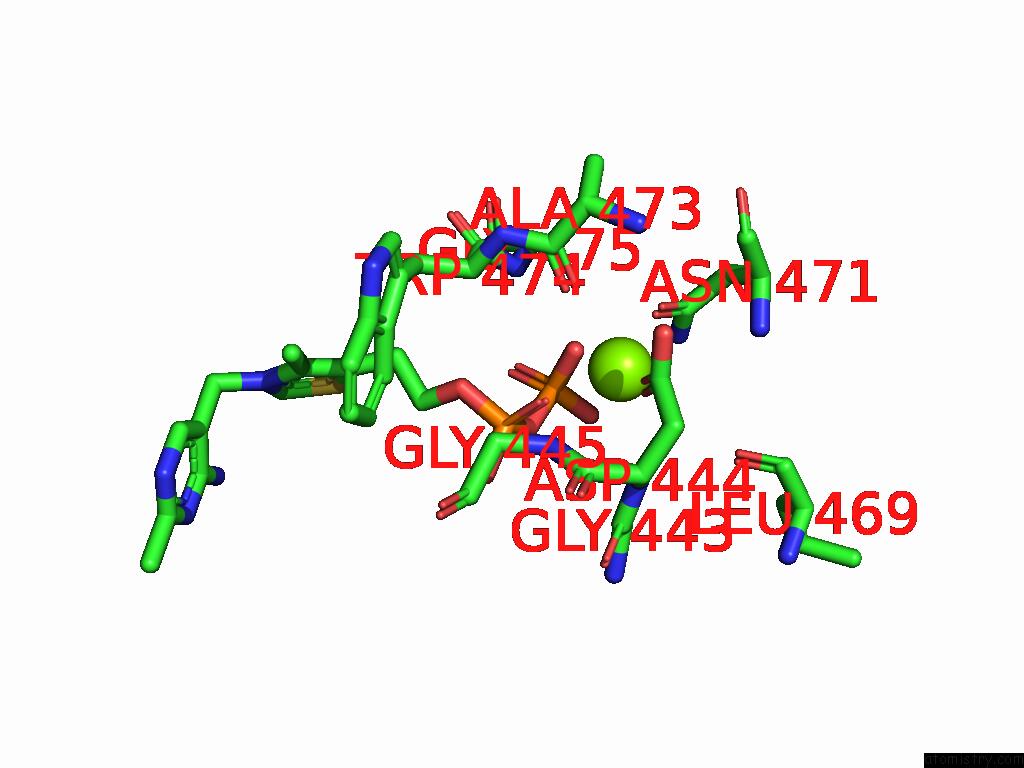
Mono view
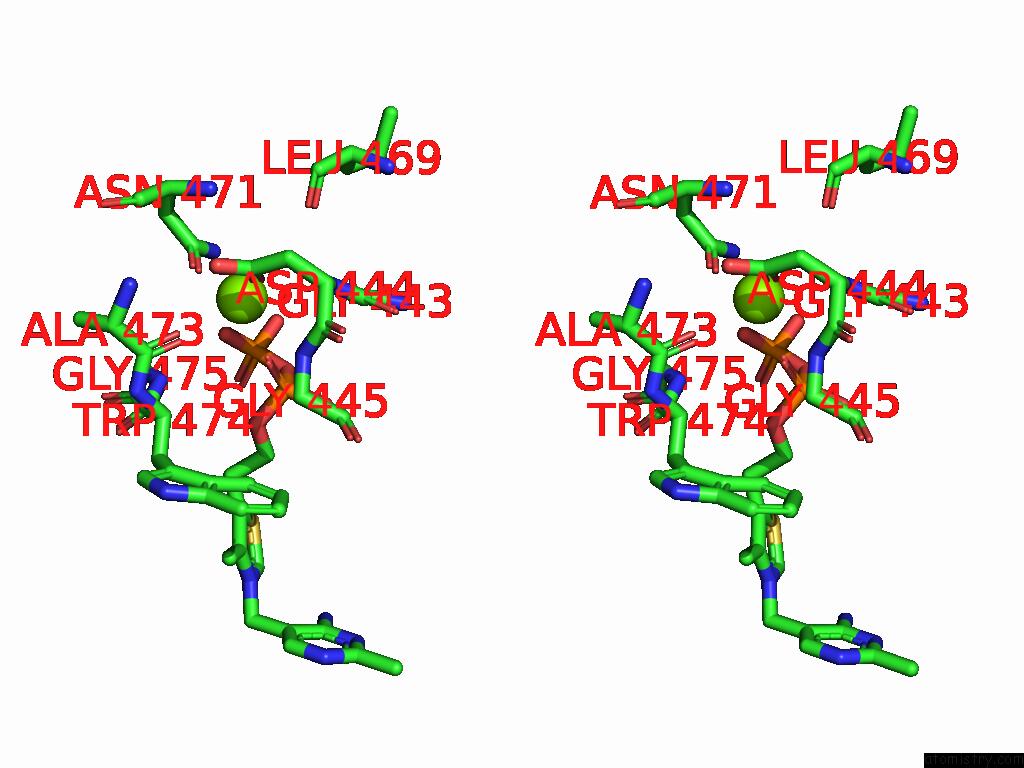
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 9 of Crystal Structure of A Benzaldehyde Lyase Mutant M6 From Herbiconiux Sp. Salv-R1 within 5.0Å range:
|
Magnesium binding site 10 out of 12 in 8y7s
Go back to
Magnesium binding site 10 out
of 12 in the Crystal Structure of A Benzaldehyde Lyase Mutant M6 From Herbiconiux Sp. Salv-R1
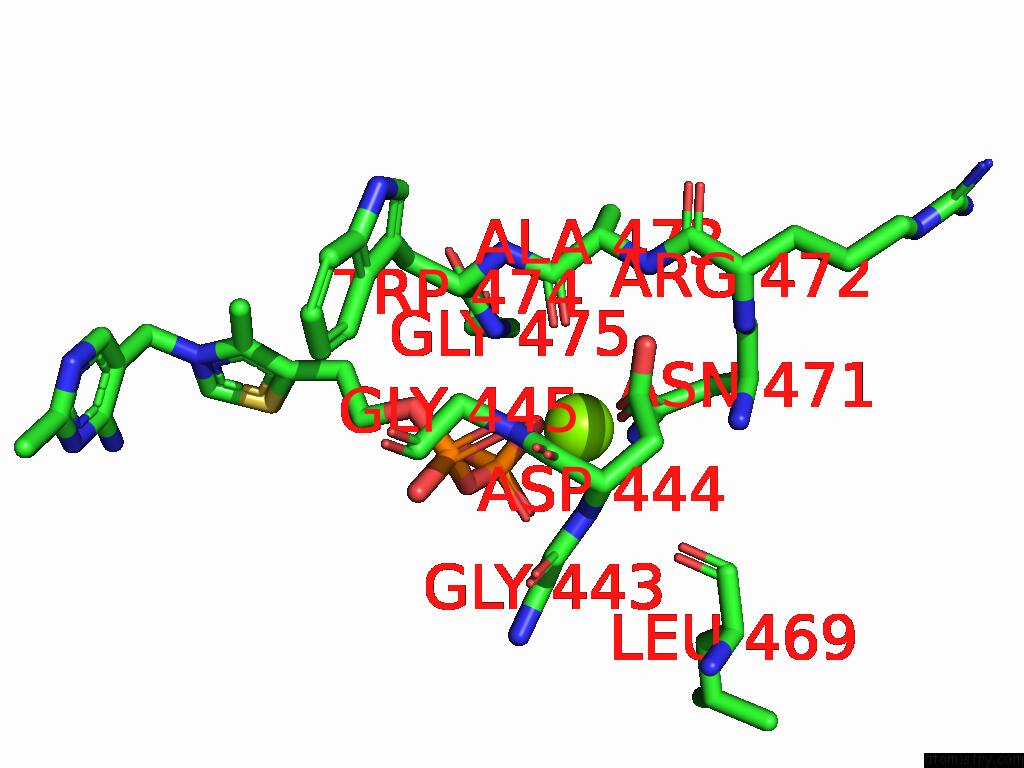
Mono view
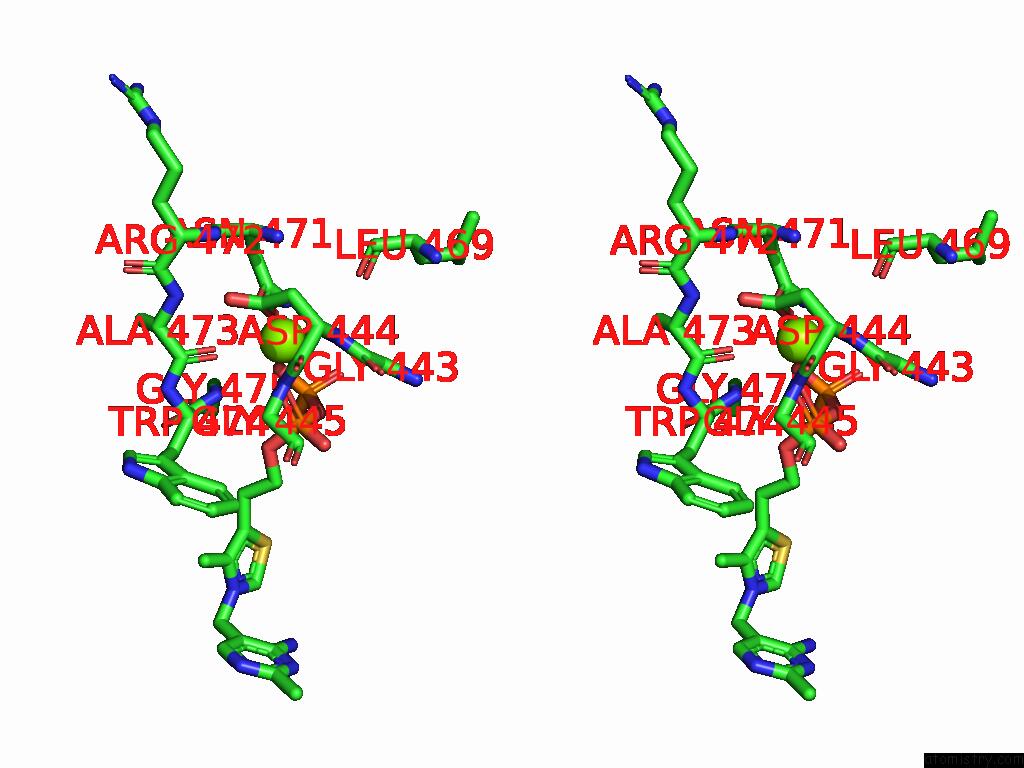
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 10 of Crystal Structure of A Benzaldehyde Lyase Mutant M6 From Herbiconiux Sp. Salv-R1 within 5.0Å range:
|
Reference:
Y.Zhang,
Y.Li,
Y.Chen,
W.Liu,
Q.Zhao,
J.Feng,
P.Yao,
Q.Wu,
D.Zhu.
Manipulating Activity and Chemoselectivity of A Benzaldehyde Lyase For Efficient Synthesis of Alpha-Hydroxymethyl Ketones and One-Pot Enantio-Complementary Conversion to 1,2-Diols Acs Catalysis V. 14 9687 2024.
ISSN: ESSN 2155-5435
DOI: 10.1021/ACSCATAL.4C01804
Page generated: Fri Aug 15 21:20:13 2025
ISSN: ESSN 2155-5435
DOI: 10.1021/ACSCATAL.4C01804
Last articles
Mn in 7EMUMn in 7E5C
Mn in 7ELV
Mn in 7ELB
Mn in 7ELC
Mn in 7EDA
Mn in 7E9W
Mn in 7ELA
Mn in 7EL9
Mn in 7DOG