Magnesium »
PDB 8zxg-9b27 »
9ash »
Magnesium in PDB 9ash: Cryo-Em Structure of the Active Lactococcus Lactis Csm Bound to Target in Post-Cleavage Stage
Magnesium Binding Sites:
The binding sites of Magnesium atom in the Cryo-Em Structure of the Active Lactococcus Lactis Csm Bound to Target in Post-Cleavage Stage
(pdb code 9ash). This binding sites where shown within
5.0 Angstroms radius around Magnesium atom.
In total 3 binding sites of Magnesium where determined in the Cryo-Em Structure of the Active Lactococcus Lactis Csm Bound to Target in Post-Cleavage Stage, PDB code: 9ash:
Jump to Magnesium binding site number: 1; 2; 3;
In total 3 binding sites of Magnesium where determined in the Cryo-Em Structure of the Active Lactococcus Lactis Csm Bound to Target in Post-Cleavage Stage, PDB code: 9ash:
Jump to Magnesium binding site number: 1; 2; 3;
Magnesium binding site 1 out of 3 in 9ash
Go back to
Magnesium binding site 1 out
of 3 in the Cryo-Em Structure of the Active Lactococcus Lactis Csm Bound to Target in Post-Cleavage Stage
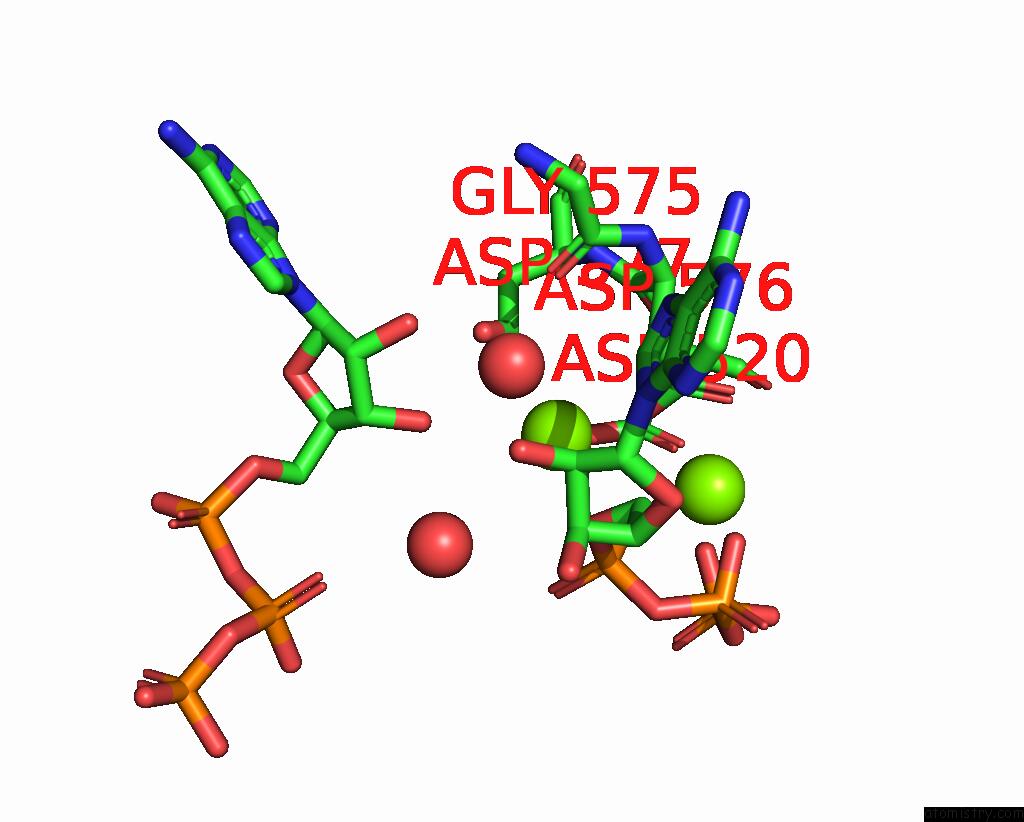
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 1 of Cryo-Em Structure of the Active Lactococcus Lactis Csm Bound to Target in Post-Cleavage Stage within 5.0Å range:
|
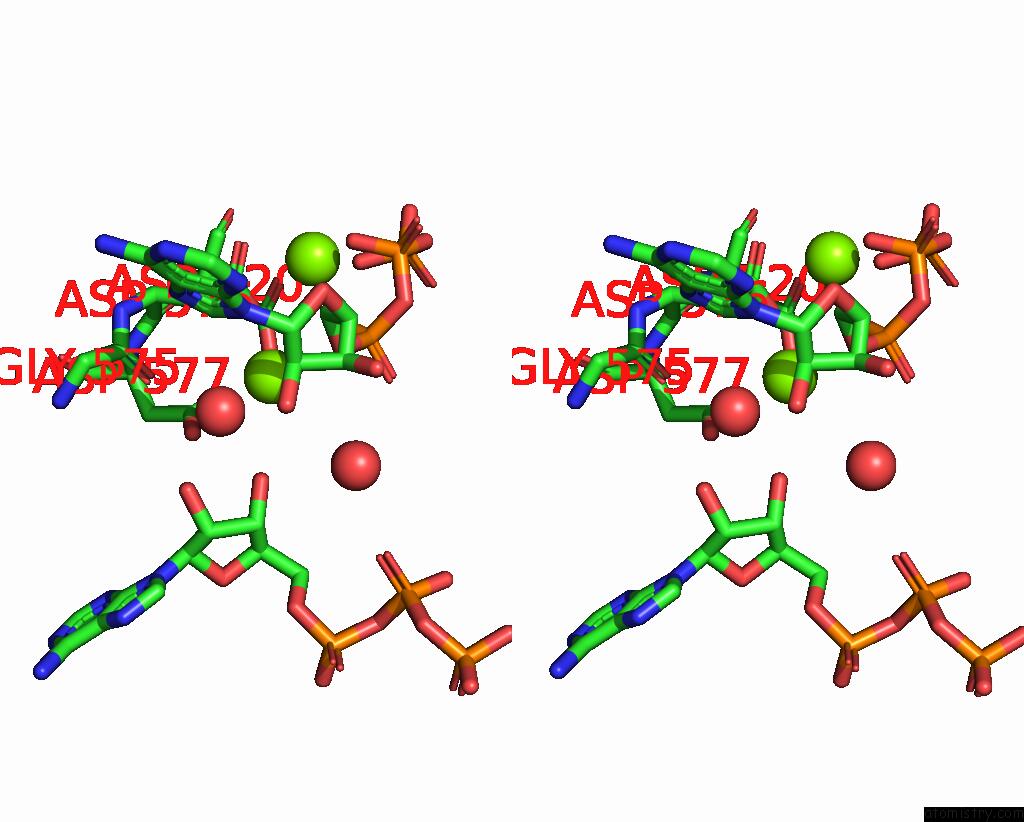
Magnesium binding site 2 out of 3 in 9ash
Go back to
Magnesium binding site 2 out
of 3 in the Cryo-Em Structure of the Active Lactococcus Lactis Csm Bound to Target in Post-Cleavage Stage
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 2 of Cryo-Em Structure of the Active Lactococcus Lactis Csm Bound to Target in Post-Cleavage Stage within 5.0Å range:
|
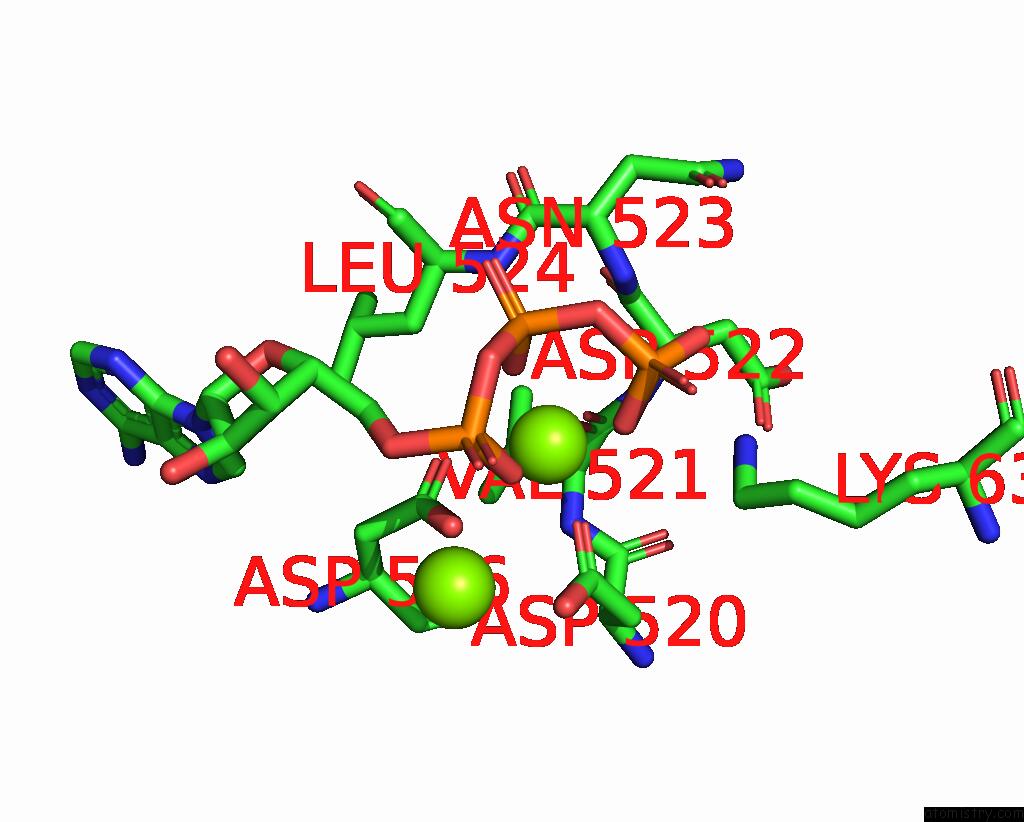
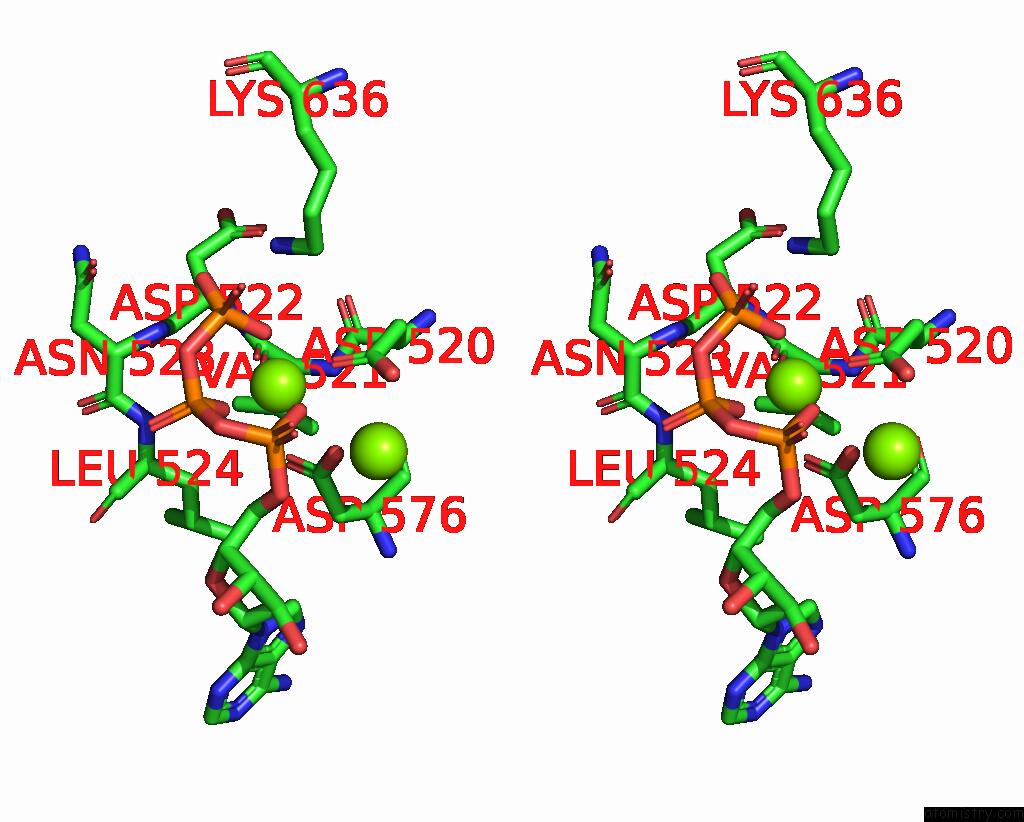
Magnesium binding site 3 out of 3 in 9ash
Go back to
Magnesium binding site 3 out
of 3 in the Cryo-Em Structure of the Active Lactococcus Lactis Csm Bound to Target in Post-Cleavage Stage
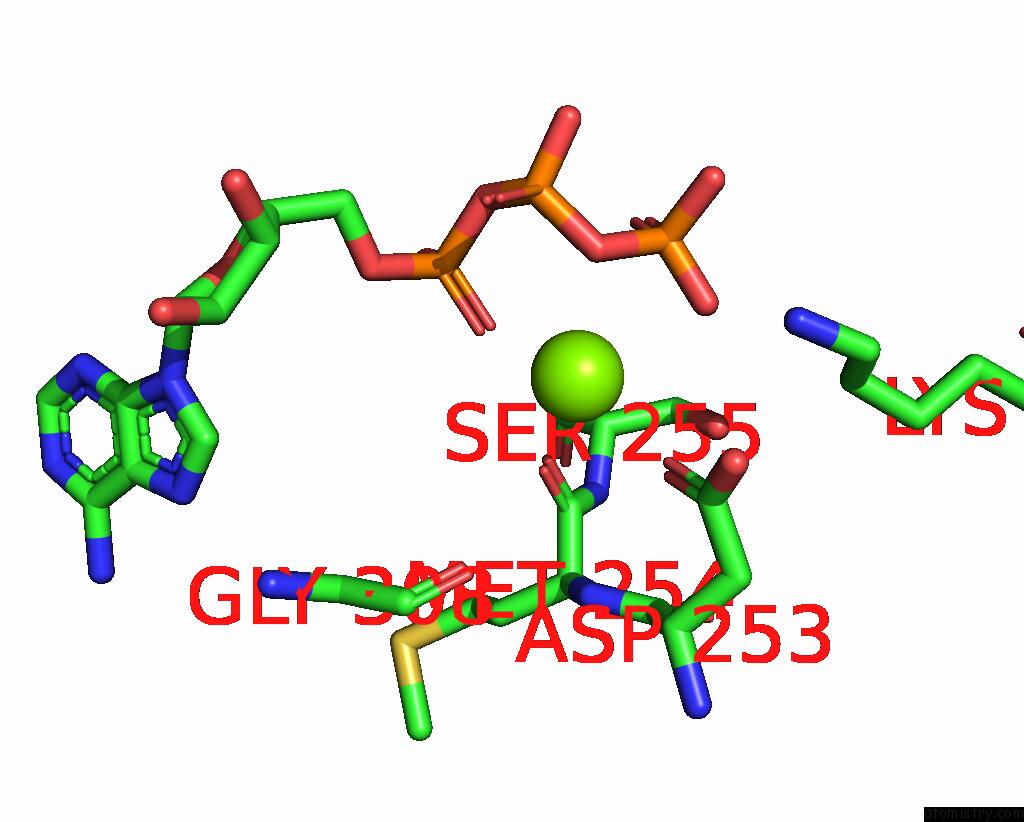
Mono view
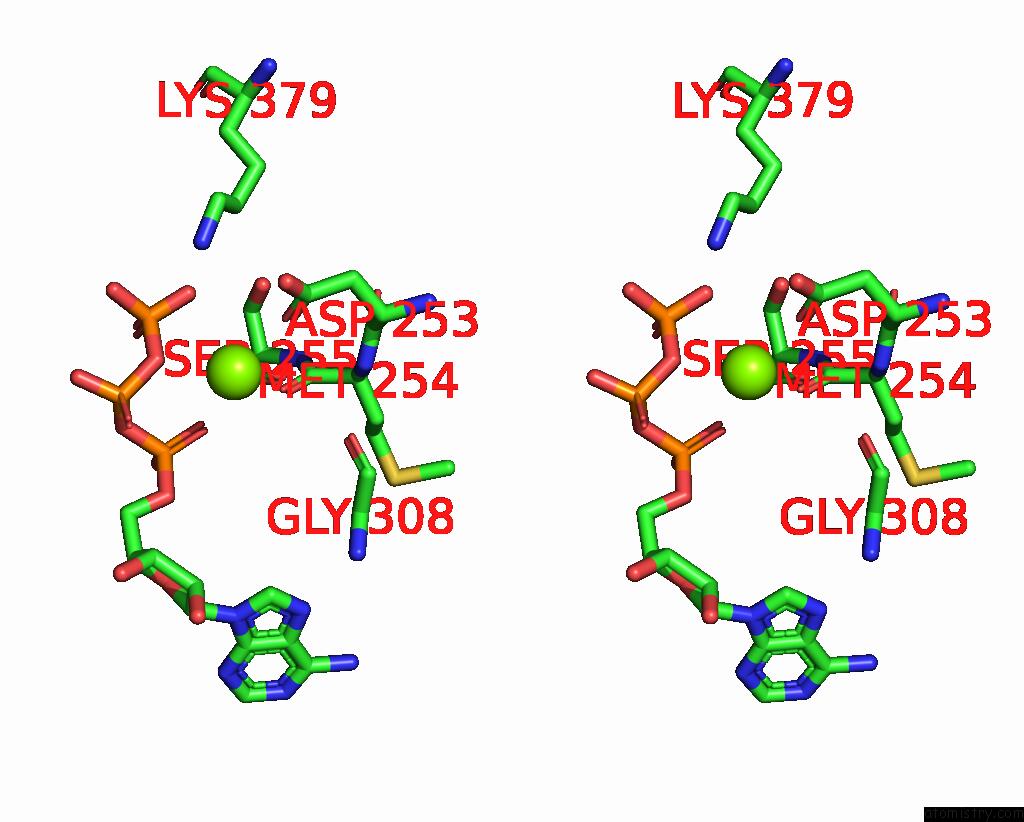
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 3 of Cryo-Em Structure of the Active Lactococcus Lactis Csm Bound to Target in Post-Cleavage Stage within 5.0Å range:
|
Reference:
H.N.Goswami,
F.Ahmadizadeh,
B.Wang,
D.Addo-Yobo,
Y.Zhao,
A.C.Whittington,
H.He,
M.P.Terns,
H.Li.
Molecular Basis For CA6 Synthesis By A Type III-A Crispr-Cas Enzyme and Its Conversion to CA4 Production. Nucleic Acids Res. 2024.
ISSN: ESSN 1362-4962
PubMed: 38989619
DOI: 10.1093/NAR/GKAE603
Page generated: Fri Aug 15 23:02:44 2025
ISSN: ESSN 1362-4962
PubMed: 38989619
DOI: 10.1093/NAR/GKAE603
Last articles
Mg in 9ED0Mg in 9EH5
Mg in 9EI1
Mg in 9EHZ
Mg in 9EFU
Mg in 9EFG
Mg in 9ED3
Mg in 9ECO
Mg in 9E9Q
Mg in 9EB5