Magnesium »
PDB 8zxg-9b27 »
9azl »
Magnesium in PDB 9azl: Cryo-Em Structure of the Abc Transporter PCAT1 Bound with Mgadp CLASS1
Magnesium Binding Sites:
The binding sites of Magnesium atom in the Cryo-Em Structure of the Abc Transporter PCAT1 Bound with Mgadp CLASS1
(pdb code 9azl). This binding sites where shown within
5.0 Angstroms radius around Magnesium atom.
In total 2 binding sites of Magnesium where determined in the Cryo-Em Structure of the Abc Transporter PCAT1 Bound with Mgadp CLASS1, PDB code: 9azl:
Jump to Magnesium binding site number: 1; 2;
In total 2 binding sites of Magnesium where determined in the Cryo-Em Structure of the Abc Transporter PCAT1 Bound with Mgadp CLASS1, PDB code: 9azl:
Jump to Magnesium binding site number: 1; 2;
Magnesium binding site 1 out of 2 in 9azl
Go back to
Magnesium binding site 1 out
of 2 in the Cryo-Em Structure of the Abc Transporter PCAT1 Bound with Mgadp CLASS1
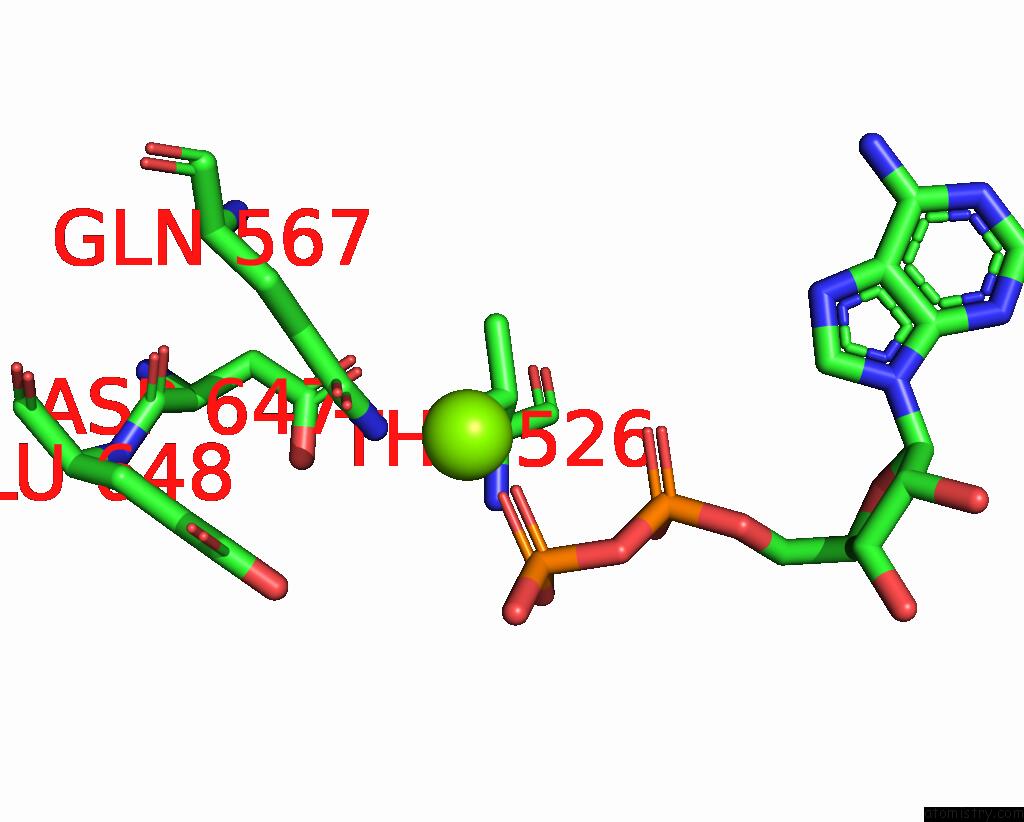
Mono view
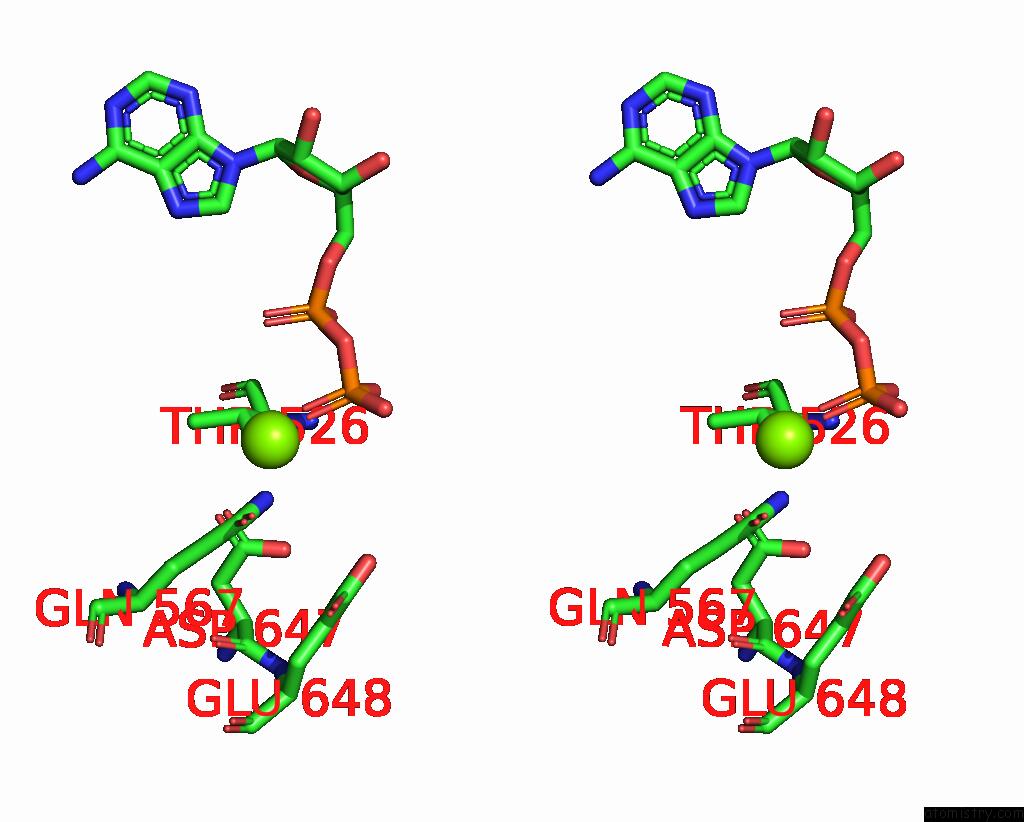
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 1 of Cryo-Em Structure of the Abc Transporter PCAT1 Bound with Mgadp CLASS1 within 5.0Å range:
|
Magnesium binding site 2 out of 2 in 9azl
Go back to
Magnesium binding site 2 out
of 2 in the Cryo-Em Structure of the Abc Transporter PCAT1 Bound with Mgadp CLASS1
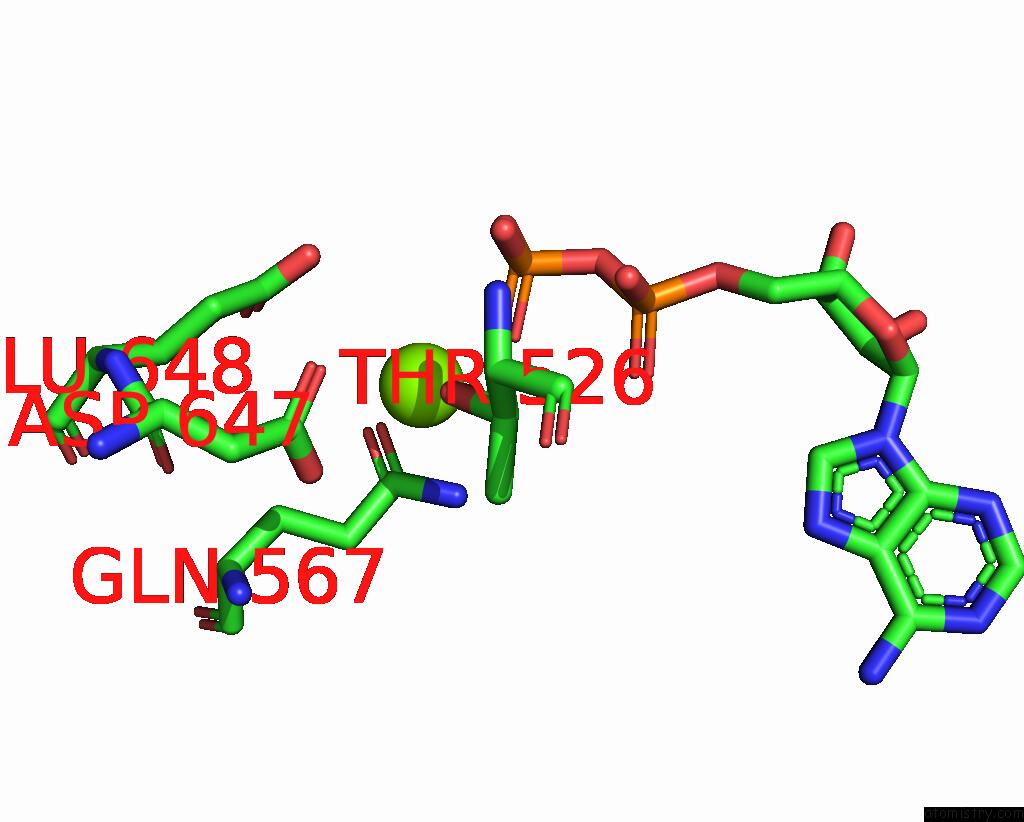
Mono view
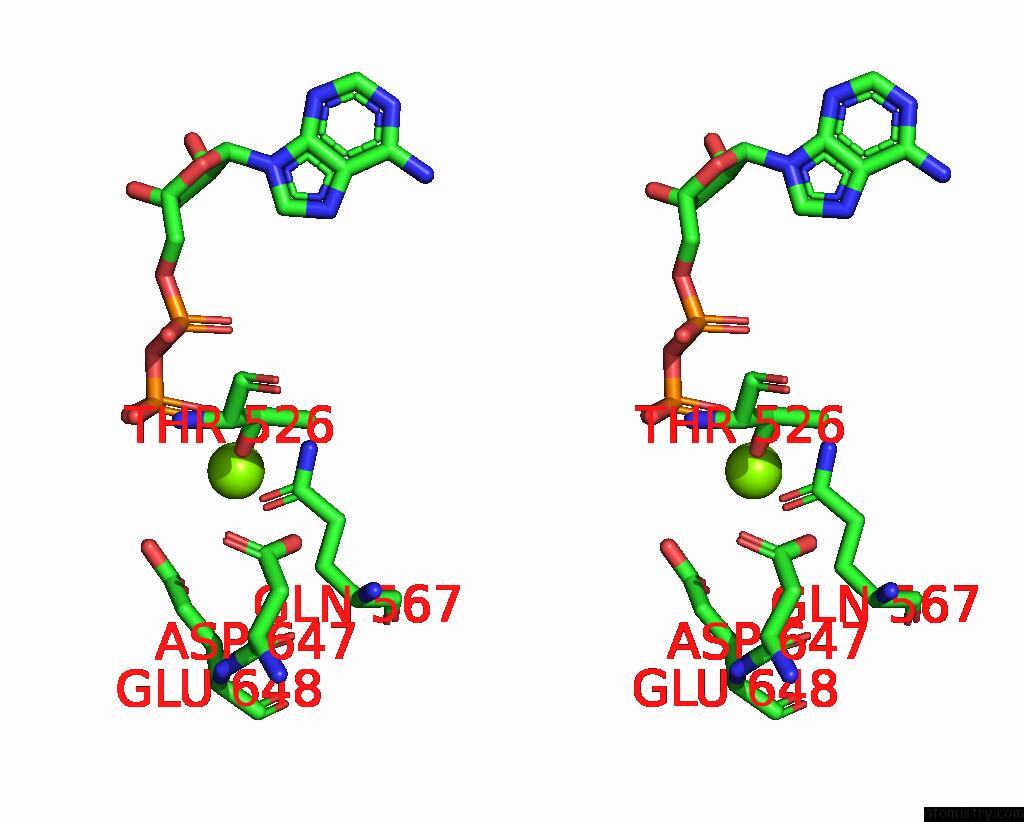
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 2 of Cryo-Em Structure of the Abc Transporter PCAT1 Bound with Mgadp CLASS1 within 5.0Å range:
|
Reference:
R.Zhang,
K.L.Jagessar,
M.Brownd,
A.Polasa,
R.A.Stein,
M.Moradi,
E.Karakas,
H.S.Mchaourab.
Conformational Cycle of A Protease-Containing Abc Transporter in Lipid Nanodiscs Reveals the Mechanism of Cargo-Protein Coupling. Nat Commun V. 15 9055 2024.
ISSN: ESSN 2041-1723
PubMed: 39428489
DOI: 10.1038/S41467-024-53420-0
Page generated: Fri Aug 15 23:07:13 2025
ISSN: ESSN 2041-1723
PubMed: 39428489
DOI: 10.1038/S41467-024-53420-0
Last articles
Mg in 9ES6Mg in 9ES9
Mg in 9ES7
Mg in 9ES2
Mg in 9ES4
Mg in 9ES0
Mg in 9ES5
Mg in 9ES1
Mg in 9ERR
Mg in 9ERV