Magnesium »
PDB 9bz9-9c87 »
9c87 »
Magnesium in PDB 9c87: Cryo-Em Structure of A Proteolytic Clpxp Aaa+ Machine Poised to Unfold A Linear-Degron Dhfr-Ssra Substrate Bound with Mtx
Enzymatic activity of Cryo-Em Structure of A Proteolytic Clpxp Aaa+ Machine Poised to Unfold A Linear-Degron Dhfr-Ssra Substrate Bound with Mtx
All present enzymatic activity of Cryo-Em Structure of A Proteolytic Clpxp Aaa+ Machine Poised to Unfold A Linear-Degron Dhfr-Ssra Substrate Bound with Mtx:
1.5.1.3; 3.4.21.92;
1.5.1.3; 3.4.21.92;
Magnesium Binding Sites:
The binding sites of Magnesium atom in the Cryo-Em Structure of A Proteolytic Clpxp Aaa+ Machine Poised to Unfold A Linear-Degron Dhfr-Ssra Substrate Bound with Mtx
(pdb code 9c87). This binding sites where shown within
5.0 Angstroms radius around Magnesium atom.
In total 4 binding sites of Magnesium where determined in the Cryo-Em Structure of A Proteolytic Clpxp Aaa+ Machine Poised to Unfold A Linear-Degron Dhfr-Ssra Substrate Bound with Mtx, PDB code: 9c87:
Jump to Magnesium binding site number: 1; 2; 3; 4;
In total 4 binding sites of Magnesium where determined in the Cryo-Em Structure of A Proteolytic Clpxp Aaa+ Machine Poised to Unfold A Linear-Degron Dhfr-Ssra Substrate Bound with Mtx, PDB code: 9c87:
Jump to Magnesium binding site number: 1; 2; 3; 4;
Magnesium binding site 1 out of 4 in 9c87
Go back to
Magnesium binding site 1 out
of 4 in the Cryo-Em Structure of A Proteolytic Clpxp Aaa+ Machine Poised to Unfold A Linear-Degron Dhfr-Ssra Substrate Bound with Mtx
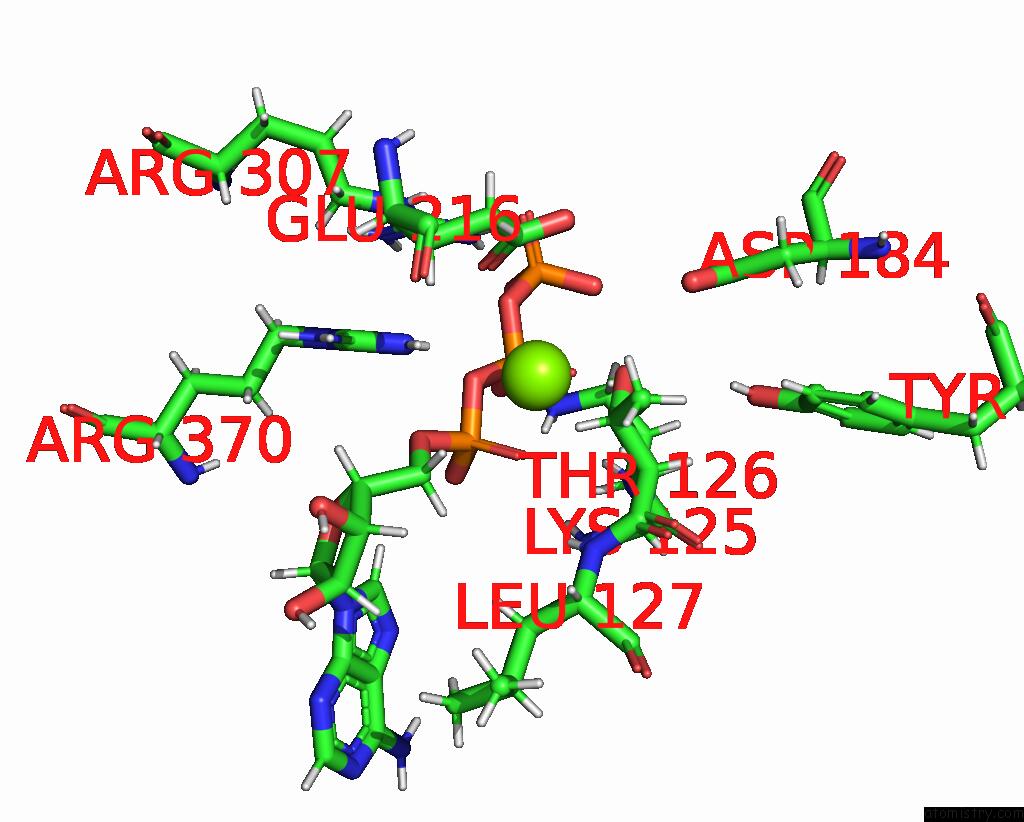
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 1 of Cryo-Em Structure of A Proteolytic Clpxp Aaa+ Machine Poised to Unfold A Linear-Degron Dhfr-Ssra Substrate Bound with Mtx within 5.0Å range:
|
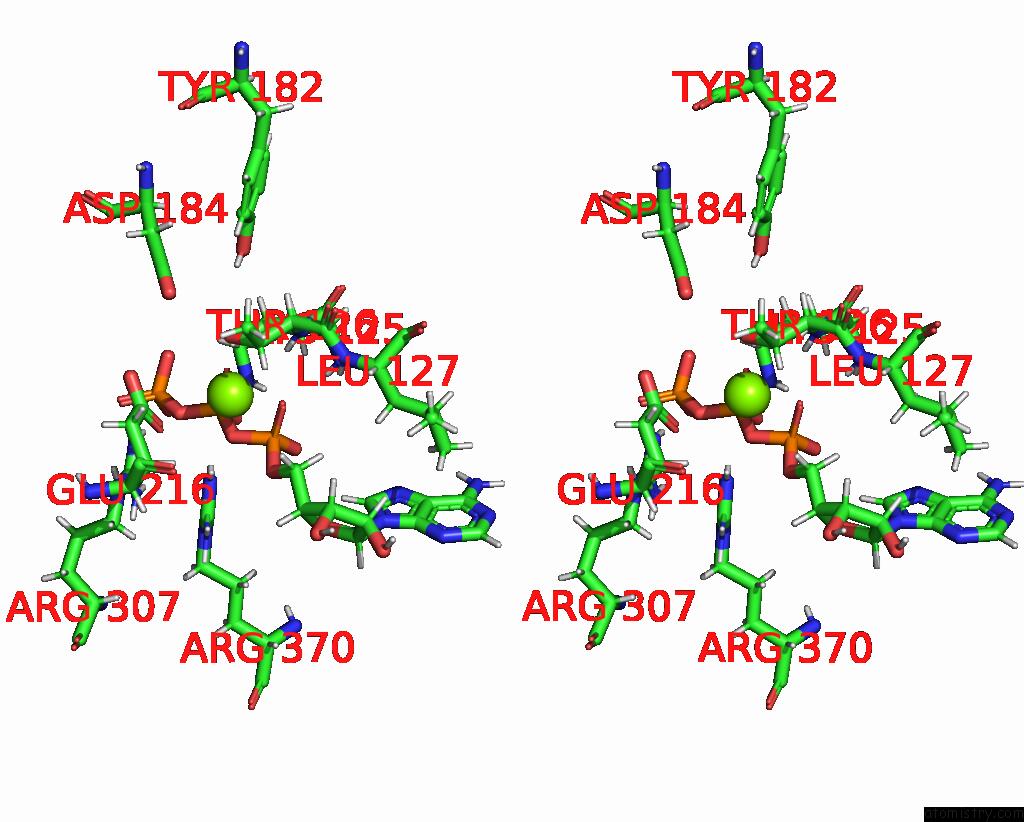
Magnesium binding site 2 out of 4 in 9c87
Go back to
Magnesium binding site 2 out
of 4 in the Cryo-Em Structure of A Proteolytic Clpxp Aaa+ Machine Poised to Unfold A Linear-Degron Dhfr-Ssra Substrate Bound with Mtx
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 2 of Cryo-Em Structure of A Proteolytic Clpxp Aaa+ Machine Poised to Unfold A Linear-Degron Dhfr-Ssra Substrate Bound with Mtx within 5.0Å range:
|
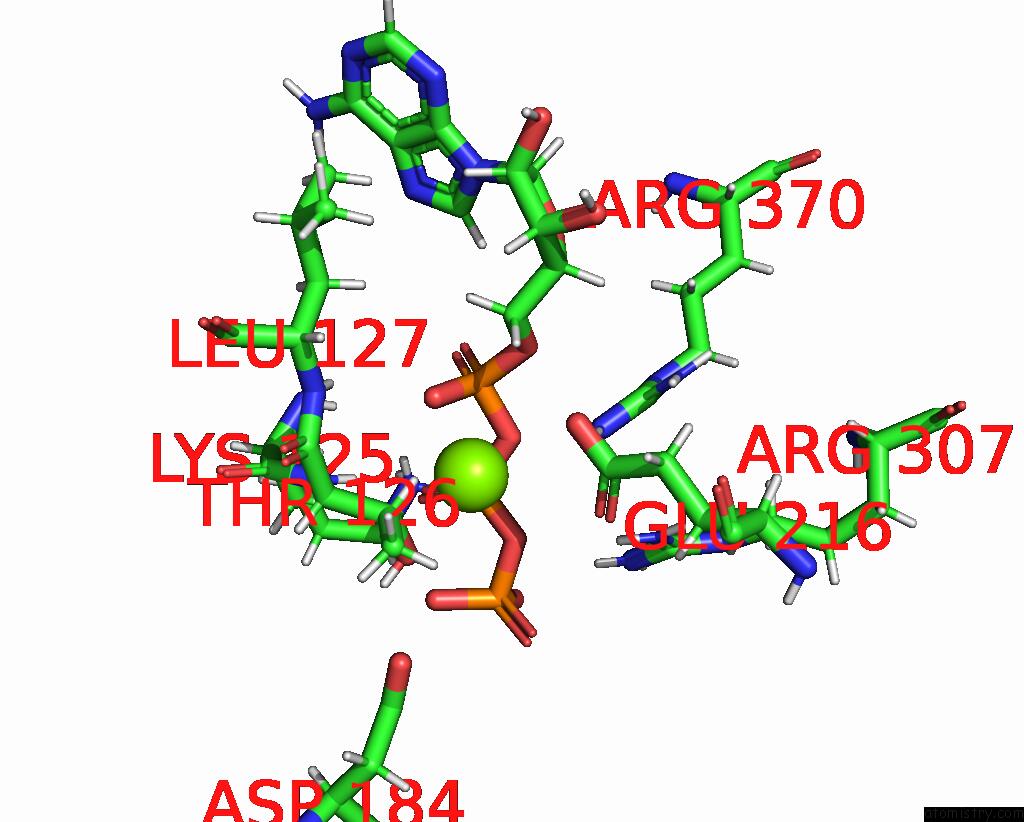
Magnesium binding site 3 out of 4 in 9c87
Go back to
Magnesium binding site 3 out
of 4 in the Cryo-Em Structure of A Proteolytic Clpxp Aaa+ Machine Poised to Unfold A Linear-Degron Dhfr-Ssra Substrate Bound with Mtx
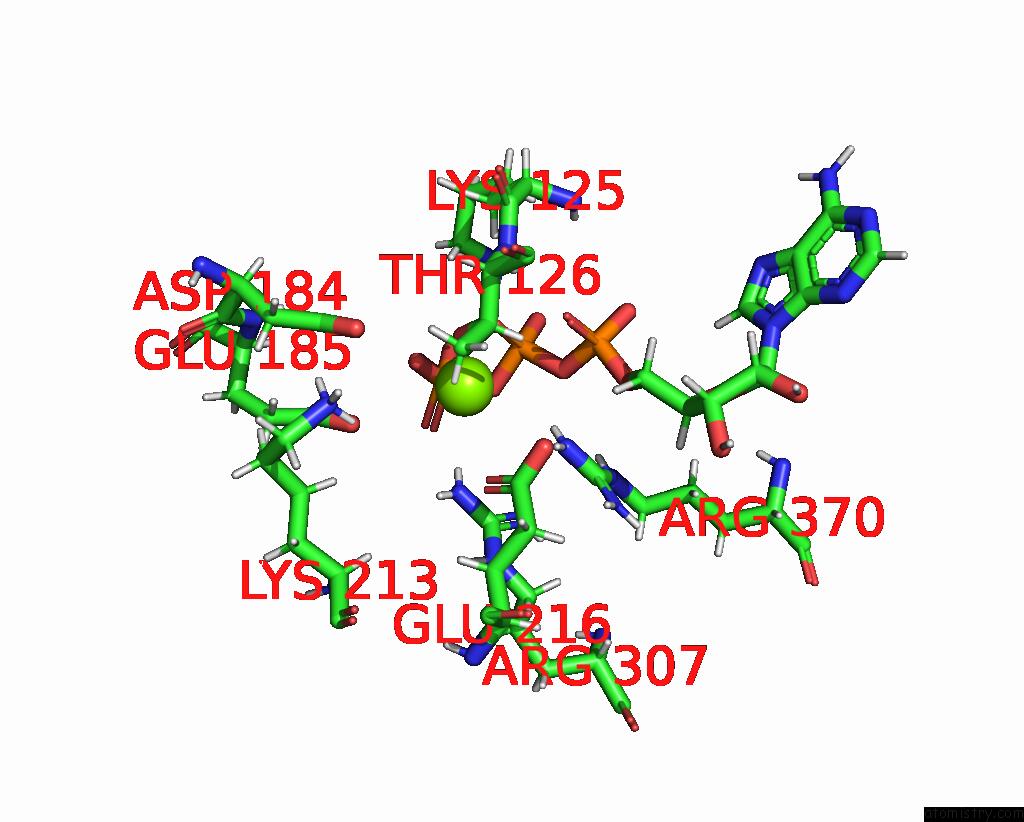
Mono view
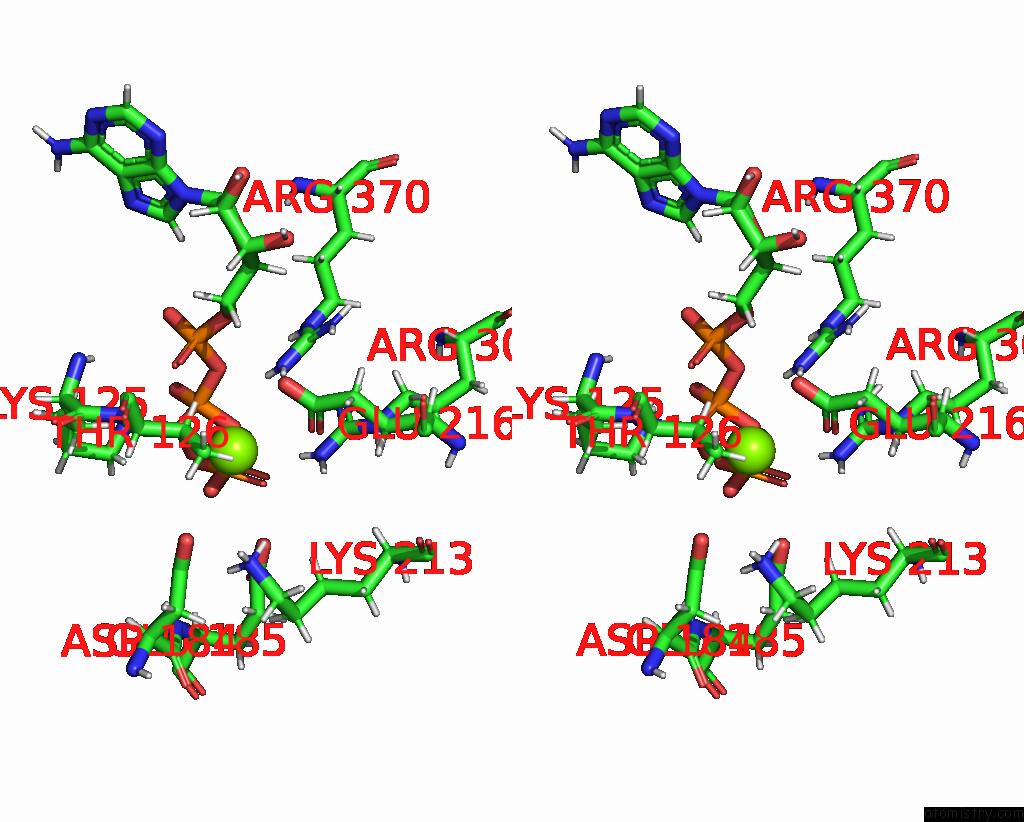
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 3 of Cryo-Em Structure of A Proteolytic Clpxp Aaa+ Machine Poised to Unfold A Linear-Degron Dhfr-Ssra Substrate Bound with Mtx within 5.0Å range:
|
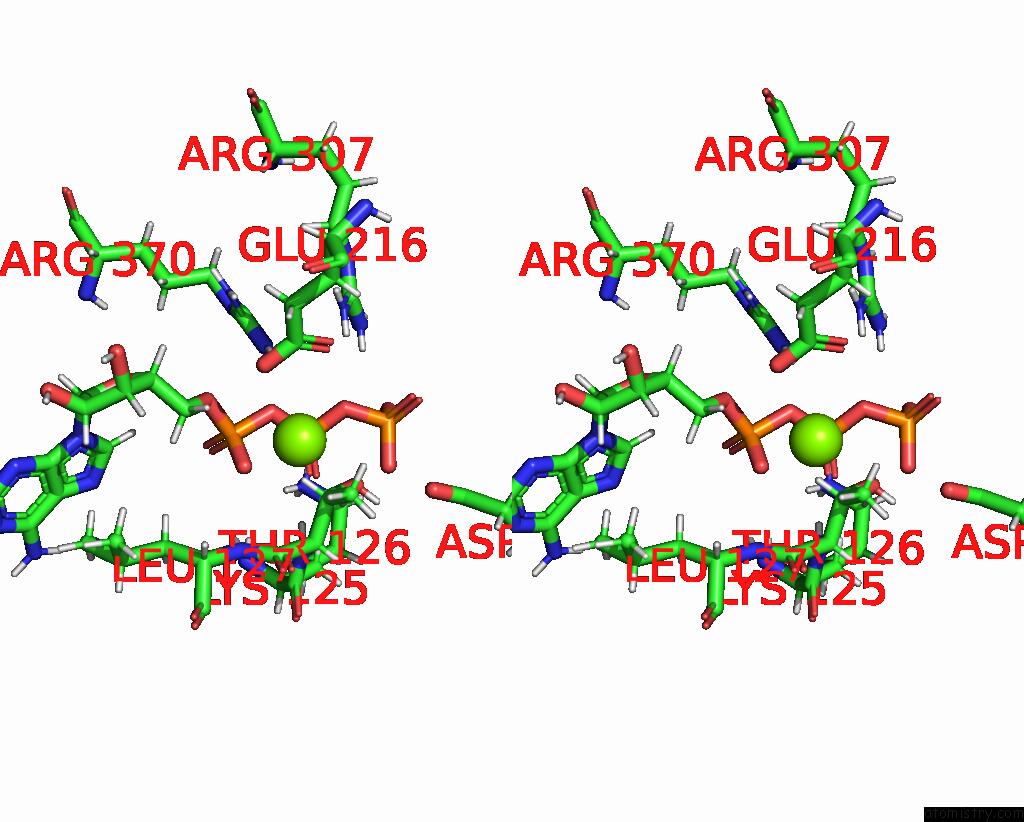
Magnesium binding site 4 out of 4 in 9c87
Go back to
Magnesium binding site 4 out
of 4 in the Cryo-Em Structure of A Proteolytic Clpxp Aaa+ Machine Poised to Unfold A Linear-Degron Dhfr-Ssra Substrate Bound with Mtx
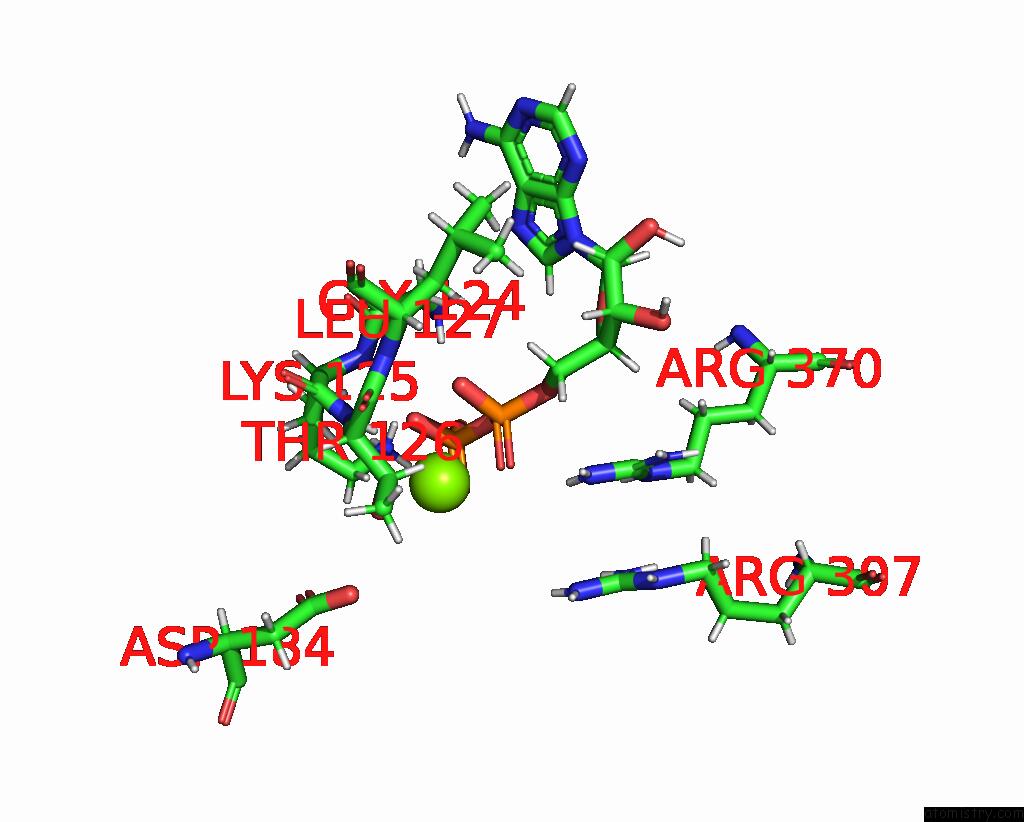
Mono view
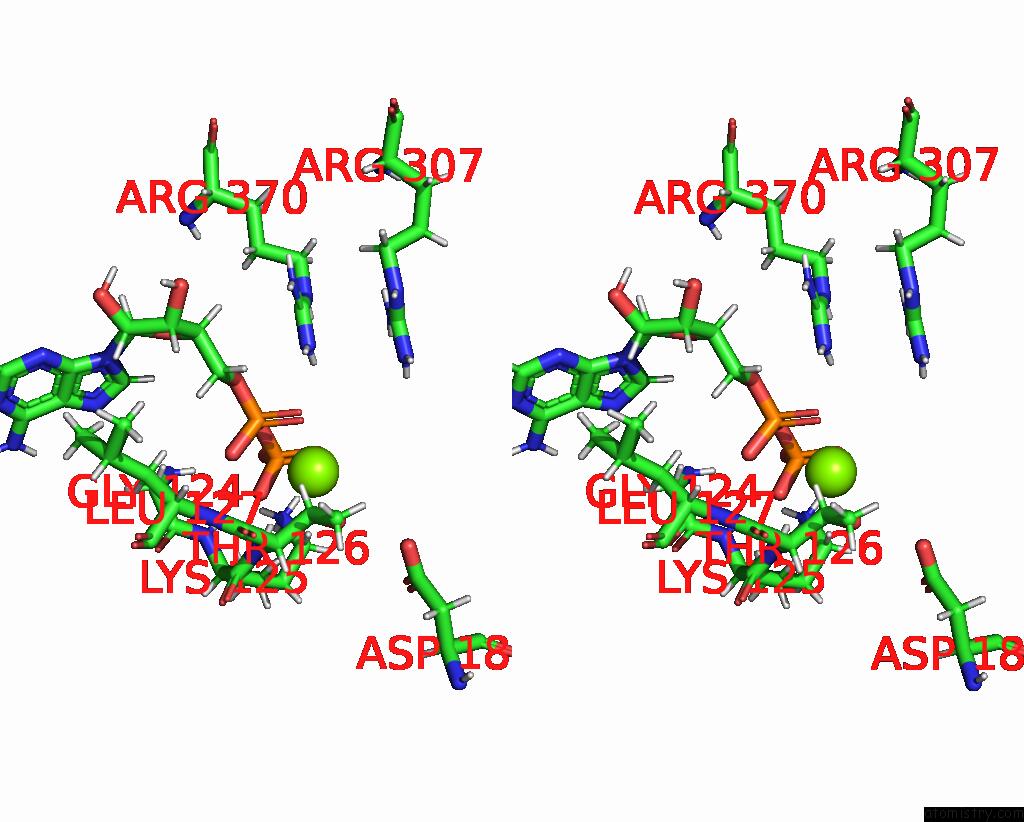
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 4 of Cryo-Em Structure of A Proteolytic Clpxp Aaa+ Machine Poised to Unfold A Linear-Degron Dhfr-Ssra Substrate Bound with Mtx within 5.0Å range:
|
Reference:
A.Ghanbarpour,
R.T.Sauer,
J.H.Davis.
Cryo-Em Structure of A Proteolytic Clpxp Aaa+ Machine Poised to Unfold A Linear-Degron Dhfr-Ssra Substrate Bound with Mtx To Be Published.
Page generated: Fri Aug 15 23:44:21 2025
Last articles
Mn in 4Q3UMn in 4Q3V
Mn in 4Q3T
Mn in 4Q3S
Mn in 4Q3Q
Mn in 4Q3R
Mn in 4Q3P
Mn in 4PXE
Mn in 4PTH
Mn in 4Q0B