Magnesium »
PDB 9mh0-9nea »
9mhh »
Magnesium in PDB 9mhh: PI3KC3-C1 in Complex with RAB1A. VPS34 Kinase Domain Active Conformation
Enzymatic activity of PI3KC3-C1 in Complex with RAB1A. VPS34 Kinase Domain Active Conformation
All present enzymatic activity of PI3KC3-C1 in Complex with RAB1A. VPS34 Kinase Domain Active Conformation:
2.7.1.137; 2.7.11.1; 3.6.5.2;
2.7.1.137; 2.7.11.1; 3.6.5.2;
Magnesium Binding Sites:
The binding sites of Magnesium atom in the PI3KC3-C1 in Complex with RAB1A. VPS34 Kinase Domain Active Conformation
(pdb code 9mhh). This binding sites where shown within
5.0 Angstroms radius around Magnesium atom.
In total 2 binding sites of Magnesium where determined in the PI3KC3-C1 in Complex with RAB1A. VPS34 Kinase Domain Active Conformation, PDB code: 9mhh:
Jump to Magnesium binding site number: 1; 2;
In total 2 binding sites of Magnesium where determined in the PI3KC3-C1 in Complex with RAB1A. VPS34 Kinase Domain Active Conformation, PDB code: 9mhh:
Jump to Magnesium binding site number: 1; 2;
Magnesium binding site 1 out of 2 in 9mhh
Go back to
Magnesium binding site 1 out
of 2 in the PI3KC3-C1 in Complex with RAB1A. VPS34 Kinase Domain Active Conformation
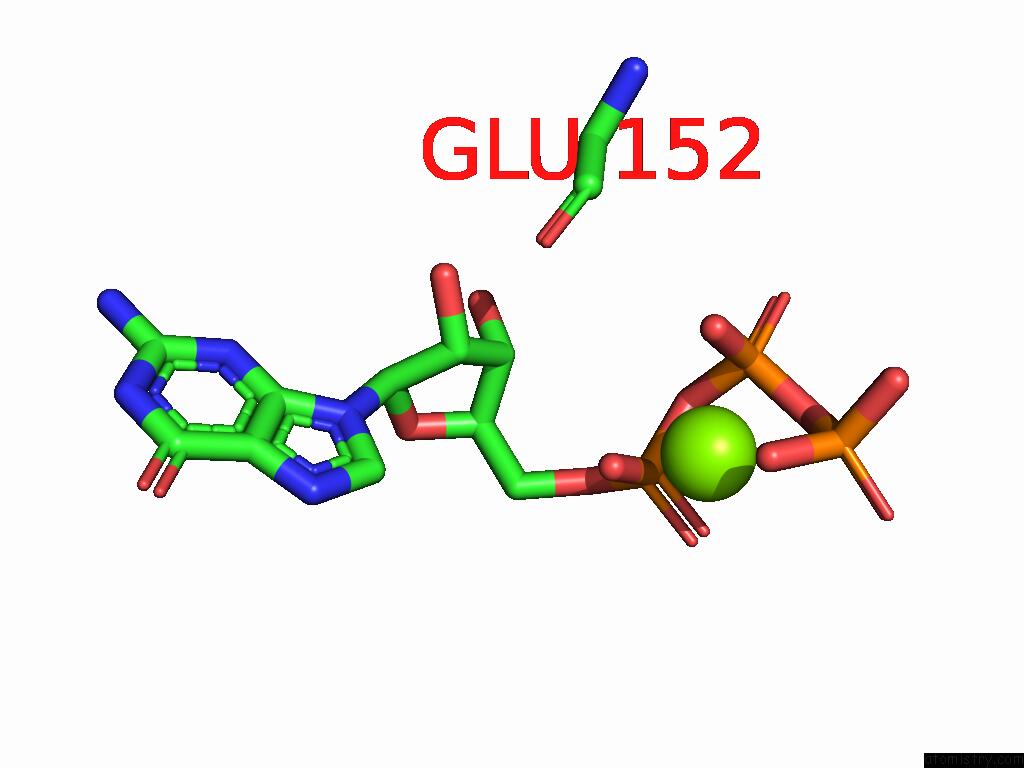
Mono view
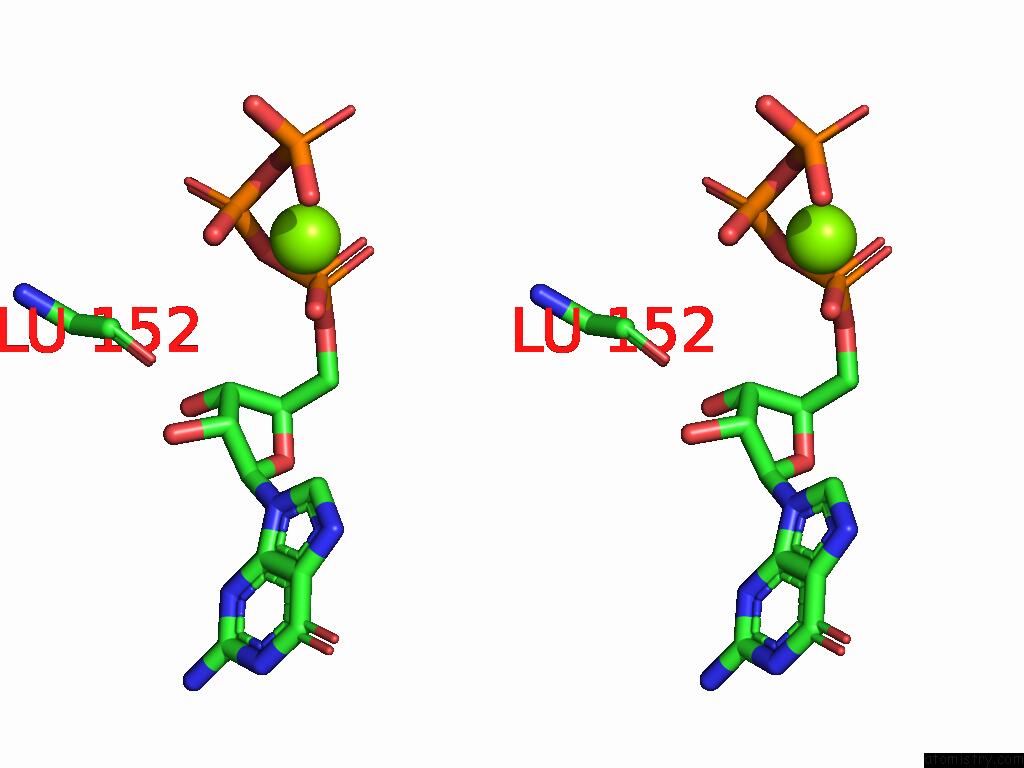
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 1 of PI3KC3-C1 in Complex with RAB1A. VPS34 Kinase Domain Active Conformation within 5.0Å range:
|
Magnesium binding site 2 out of 2 in 9mhh
Go back to
Magnesium binding site 2 out
of 2 in the PI3KC3-C1 in Complex with RAB1A. VPS34 Kinase Domain Active Conformation
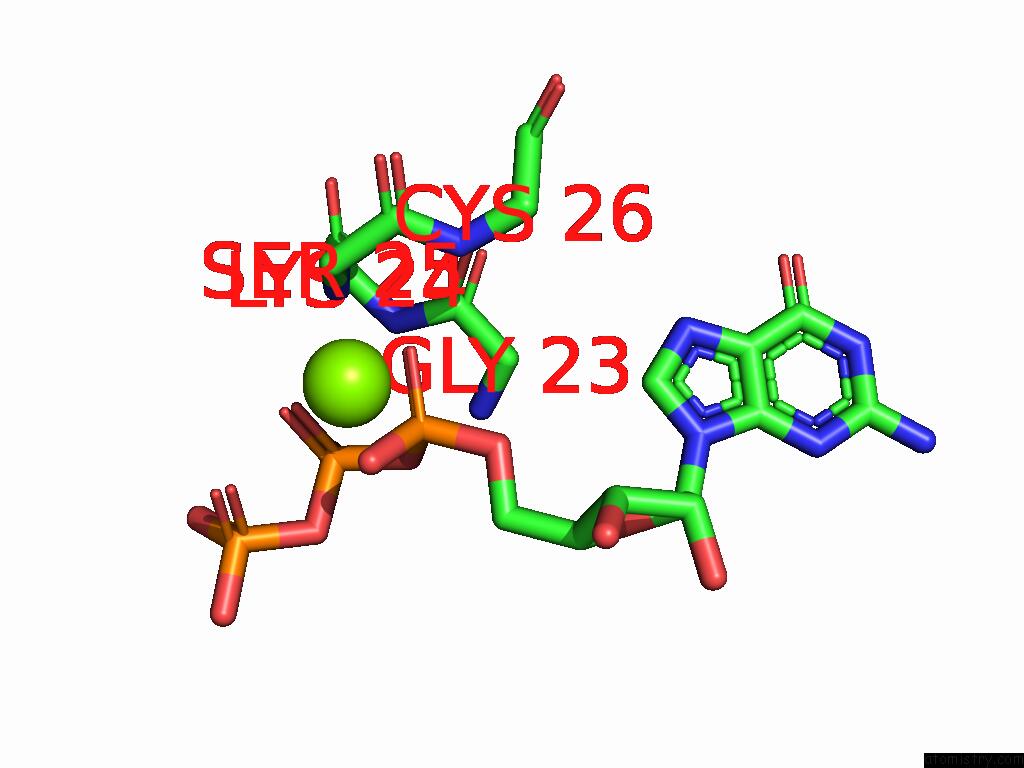
Mono view
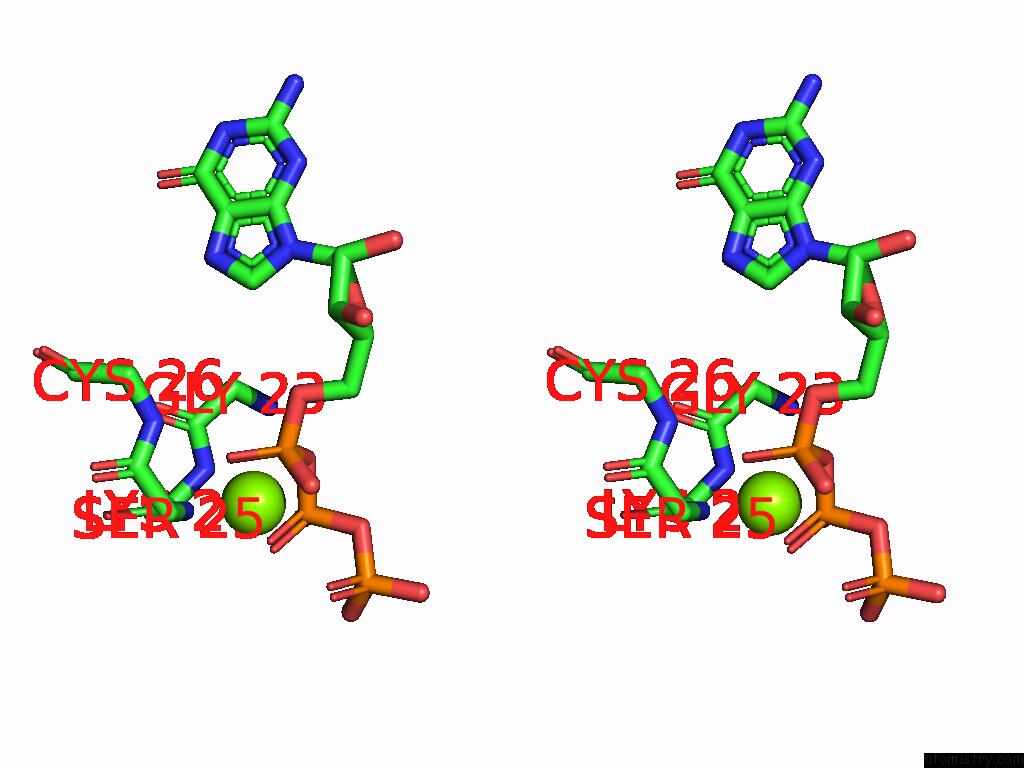
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 2 of PI3KC3-C1 in Complex with RAB1A. VPS34 Kinase Domain Active Conformation within 5.0Å range:
|
Reference:
A.S.I.Cook,
M.Chen,
T.N.Ngyuen,
A.Claveras-Cabezudo,
G.Khuu,
S.Rao,
S.N.Garcia,
M.Yang,
A.T.Iavarone,
X.Ren,
M.Lazarou,
G.Hummer,
J.H.Hurley.
Structural Pathway For Class III Pi 3-Kinase Activation By the Myristoylated Gtpbinding Pseudokinase VPS15 To Be Published.
Page generated: Sat Aug 16 06:25:08 2025
Last articles
Mn in 5FV9Mn in 5FPV
Mn in 5FXV
Mn in 5FX8
Mn in 5FWJ
Mn in 5FUP
Mn in 5FV3
Mn in 5FUN
Mn in 5FNO
Mn in 5FPL