Magnesium »
PDB 1mum-1n6i »
1n57 »
Magnesium in PDB 1n57: Crystal Structure of Chaperone HSP31
Protein crystallography data
The structure of Crystal Structure of Chaperone HSP31, PDB code: 1n57
was solved by
P.M.Quigley,
K.Korotkov,
F.Baneyx,
W.G.J.Hol,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 19.73 / 1.60 |
| Space group | C 1 2 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 52.150, 82.000, 64.480, 90.00, 100.00, 90.00 |
| R / Rfree (%) | 18.7 / 24.2 |
Magnesium Binding Sites:
The binding sites of Magnesium atom in the Crystal Structure of Chaperone HSP31
(pdb code 1n57). This binding sites where shown within
5.0 Angstroms radius around Magnesium atom.
In total only one binding site of Magnesium was determined in the Crystal Structure of Chaperone HSP31, PDB code: 1n57:
In total only one binding site of Magnesium was determined in the Crystal Structure of Chaperone HSP31, PDB code: 1n57:
Magnesium binding site 1 out of 1 in 1n57
Go back to
Magnesium binding site 1 out
of 1 in the Crystal Structure of Chaperone HSP31
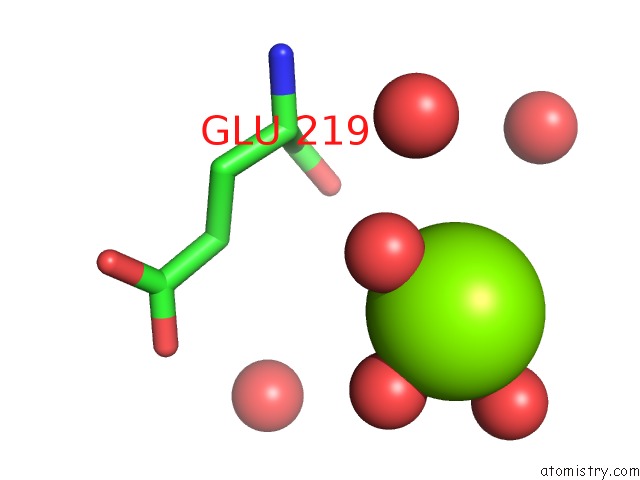
Mono view
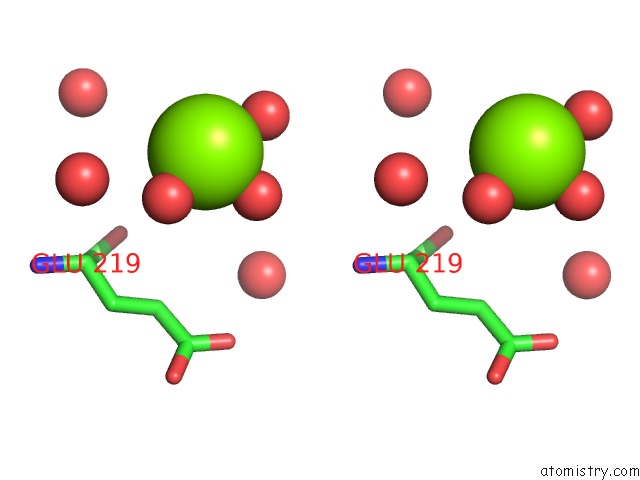
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 1 of Crystal Structure of Chaperone HSP31 within 5.0Å range:
|
Reference:
P.M.Quigley,
K.Korotkov,
F.Baneyx,
W.G.J.Hol.
The 1.6A Crystal Structure of the Class of Chaperone Represented By Escherichia Coli HSP31 Reveals A Putative Catalytic Triad Proc.Natl.Acad.Sci.Usa V. 100 3137 2003.
ISSN: ISSN 0027-8424
PubMed: 12621151
DOI: 10.1073/PNAS.0530312100
Page generated: Tue Aug 13 09:22:04 2024
ISSN: ISSN 0027-8424
PubMed: 12621151
DOI: 10.1073/PNAS.0530312100
Last articles
Br in 4CJWBr in 4CHZ
Br in 4BWO
Br in 4C8U
Br in 4C7B
Br in 4C78
Br in 4C5D
Br in 4C46
Br in 4C37
Br in 4BY7