Magnesium »
PDB 1x1t-1xfx »
1xbx »
Magnesium in PDB 1xbx: Structure of 3-Keto-L-Gulonate 6-Phosphate Decarboxylase E112D/R139V/T169A Mutant with Bound D-Ribulose 5-Phosphate
Protein crystallography data
The structure of Structure of 3-Keto-L-Gulonate 6-Phosphate Decarboxylase E112D/R139V/T169A Mutant with Bound D-Ribulose 5-Phosphate, PDB code: 1xbx
was solved by
E.L.Wise,
W.S.Yew,
J.Akana,
J.A.Gerlt,
I.Rayment,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 90.00 / 1.81 |
| Space group | C 1 2 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 123.000, 41.831, 91.068, 90.00, 97.47, 90.00 |
| R / Rfree (%) | 14.1 / 17.8 |
Magnesium Binding Sites:
The binding sites of Magnesium atom in the Structure of 3-Keto-L-Gulonate 6-Phosphate Decarboxylase E112D/R139V/T169A Mutant with Bound D-Ribulose 5-Phosphate
(pdb code 1xbx). This binding sites where shown within
5.0 Angstroms radius around Magnesium atom.
In total 2 binding sites of Magnesium where determined in the Structure of 3-Keto-L-Gulonate 6-Phosphate Decarboxylase E112D/R139V/T169A Mutant with Bound D-Ribulose 5-Phosphate, PDB code: 1xbx:
Jump to Magnesium binding site number: 1; 2;
In total 2 binding sites of Magnesium where determined in the Structure of 3-Keto-L-Gulonate 6-Phosphate Decarboxylase E112D/R139V/T169A Mutant with Bound D-Ribulose 5-Phosphate, PDB code: 1xbx:
Jump to Magnesium binding site number: 1; 2;
Magnesium binding site 1 out of 2 in 1xbx
Go back to
Magnesium binding site 1 out
of 2 in the Structure of 3-Keto-L-Gulonate 6-Phosphate Decarboxylase E112D/R139V/T169A Mutant with Bound D-Ribulose 5-Phosphate
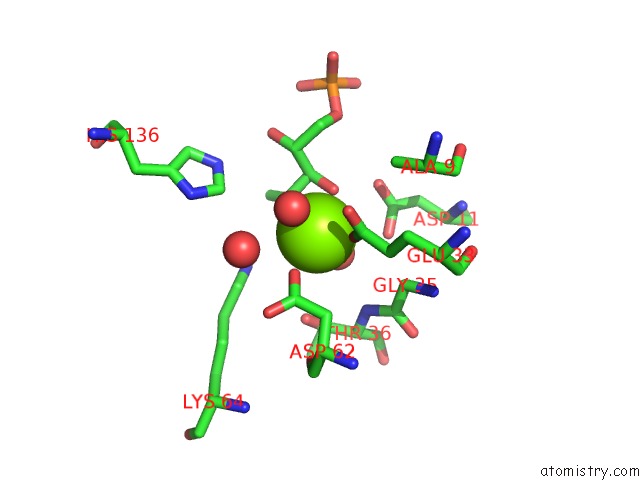
Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 1 of Structure of 3-Keto-L-Gulonate 6-Phosphate Decarboxylase E112D/R139V/T169A Mutant with Bound D-Ribulose 5-Phosphate within 5.0Å range:
|
Magnesium binding site 2 out of 2 in 1xbx
Go back to
Magnesium binding site 2 out
of 2 in the Structure of 3-Keto-L-Gulonate 6-Phosphate Decarboxylase E112D/R139V/T169A Mutant with Bound D-Ribulose 5-Phosphate

Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 2 of Structure of 3-Keto-L-Gulonate 6-Phosphate Decarboxylase E112D/R139V/T169A Mutant with Bound D-Ribulose 5-Phosphate within 5.0Å range:
|
Reference:
E.L.Wise,
W.S.Yew,
J.Akana,
J.A.Gerlt,
I.Rayment.
Evolution of Enzymatic Activities in the Orotidine 5'-Monophosphate Decarboxylase Suprafamily: Structural Basis For Catalytic Promiscuity in Wild-Type and Designed Mutants of 3-Keto-L-Gulonate 6-Phosphate Decarboxylase Biochemistry V. 44 1816 2005.
ISSN: ISSN 0006-2960
PubMed: 15697207
DOI: 10.1021/BI0478143
Page generated: Tue Aug 13 17:29:35 2024
ISSN: ISSN 0006-2960
PubMed: 15697207
DOI: 10.1021/BI0478143
Last articles
K in 3B0MK in 3B0L
K in 3B0G
K in 3B0J
K in 3AY9
K in 3AHS
K in 3AOP
K in 3ATV
K in 3AFJ
K in 3ACT