Magnesium »
PDB 208d-2a6e »
2a33 »
Magnesium in PDB 2a33: X-Ray Structure of A Lysine Decarboxylase-Like Protein From Arabidopsis Thaliana Gene AT2G37210
Protein crystallography data
The structure of X-Ray Structure of A Lysine Decarboxylase-Like Protein From Arabidopsis Thaliana Gene AT2G37210, PDB code: 2a33
was solved by
G.E.Wesenberg,
G.N.Phillips Jr.,
J.G.Mccoy,
E.Bitto,
C.A.Bingman,
S.T.M.Allard,
Center For Eukaryotic Structural Genomics (Cesg),
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 46.98 / 1.95 |
| Space group | P 21 21 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 53.403, 66.773, 98.639, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 18.1 / 23.4 |
Magnesium Binding Sites:
The binding sites of Magnesium atom in the X-Ray Structure of A Lysine Decarboxylase-Like Protein From Arabidopsis Thaliana Gene AT2G37210
(pdb code 2a33). This binding sites where shown within
5.0 Angstroms radius around Magnesium atom.
In total only one binding site of Magnesium was determined in the X-Ray Structure of A Lysine Decarboxylase-Like Protein From Arabidopsis Thaliana Gene AT2G37210, PDB code: 2a33:
In total only one binding site of Magnesium was determined in the X-Ray Structure of A Lysine Decarboxylase-Like Protein From Arabidopsis Thaliana Gene AT2G37210, PDB code: 2a33:
Magnesium binding site 1 out of 1 in 2a33
Go back to
Magnesium binding site 1 out
of 1 in the X-Ray Structure of A Lysine Decarboxylase-Like Protein From Arabidopsis Thaliana Gene AT2G37210
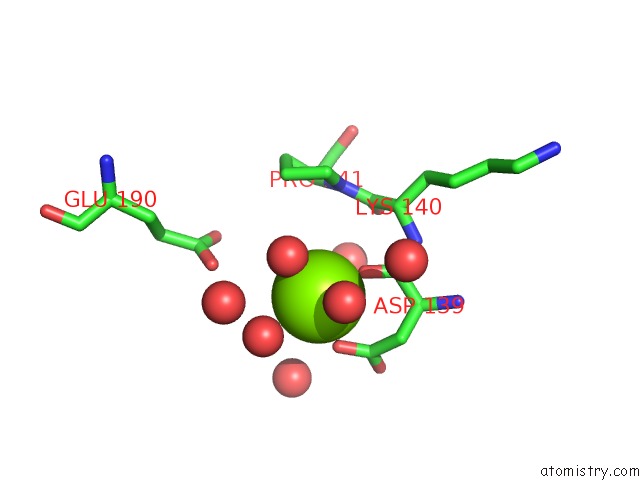
Mono view
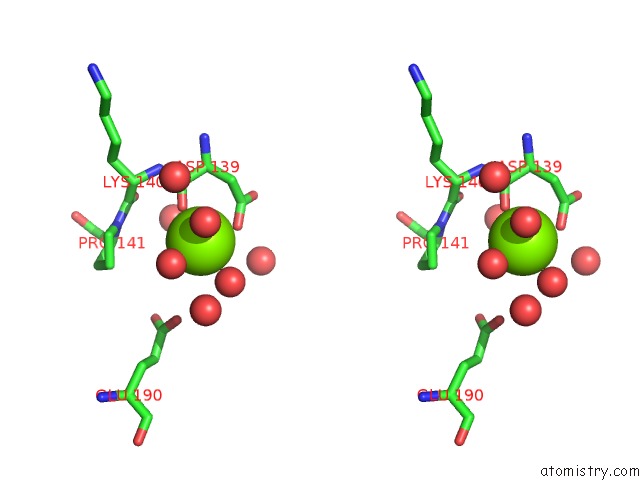
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 1 of X-Ray Structure of A Lysine Decarboxylase-Like Protein From Arabidopsis Thaliana Gene AT2G37210 within 5.0Å range:
|
Reference:
W.B.Jeon,
S.T.M.Allard,
C.A.Bingman,
E.Bitto,
B.W.Han,
G.E.Wesenberg,
G.N.Phillips Jr..
X-Ray Crystal Structures of the Conserved Hypothetical Proteins From Arabidopsis Thaliana Gene Loci AT5G11950 and AT2G37210. Proteins V. 65 1051 2006.
ISSN: ISSN 0887-3585
PubMed: 17048257
DOI: 10.1002/PROT.21166
Page generated: Sun Aug 10 09:00:36 2025
ISSN: ISSN 0887-3585
PubMed: 17048257
DOI: 10.1002/PROT.21166
Last articles
Mg in 5JVHMg in 5K0N
Mg in 5K0L
Mg in 5JZQ
Mg in 5K0K
Mg in 5JYG
Mg in 5JZJ
Mg in 5JZD
Mg in 5JYS
Mg in 5JYD