Magnesium »
PDB 3jay-3jwr »
3jva »
Magnesium in PDB 3jva: Crystal Structure of Dipeptide Epimerase From Enterococcus Faecalis V583
Protein crystallography data
The structure of Crystal Structure of Dipeptide Epimerase From Enterococcus Faecalis V583, PDB code: 3jva
was solved by
A.A.Fedorov,
E.V.Fedorov,
A.Sakai,
H.Imker,
J.A.Gerlt,
S.C.Almo,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 24.93 / 1.70 |
| Space group | H 3 |
| Cell size a, b, c (Å), α, β, γ (°) | 162.268, 162.268, 319.561, 90.00, 90.00, 120.00 |
| R / Rfree (%) | 22.3 / 24.4 |
Magnesium Binding Sites:
The binding sites of Magnesium atom in the Crystal Structure of Dipeptide Epimerase From Enterococcus Faecalis V583
(pdb code 3jva). This binding sites where shown within
5.0 Angstroms radius around Magnesium atom.
In total 8 binding sites of Magnesium where determined in the Crystal Structure of Dipeptide Epimerase From Enterococcus Faecalis V583, PDB code: 3jva:
Jump to Magnesium binding site number: 1; 2; 3; 4; 5; 6; 7; 8;
In total 8 binding sites of Magnesium where determined in the Crystal Structure of Dipeptide Epimerase From Enterococcus Faecalis V583, PDB code: 3jva:
Jump to Magnesium binding site number: 1; 2; 3; 4; 5; 6; 7; 8;
Magnesium binding site 1 out of 8 in 3jva
Go back to
Magnesium binding site 1 out
of 8 in the Crystal Structure of Dipeptide Epimerase From Enterococcus Faecalis V583
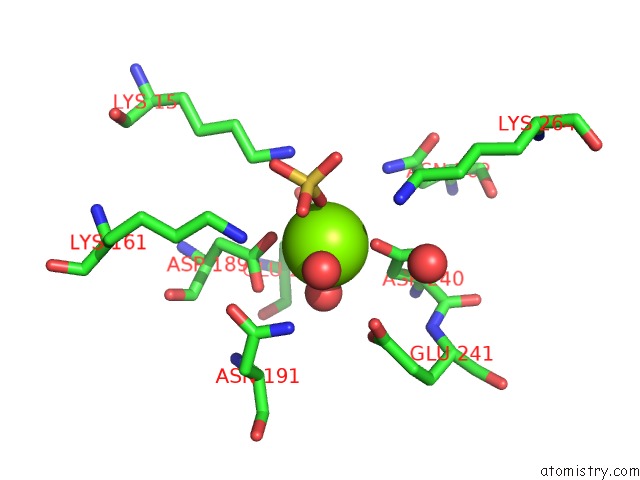
Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 1 of Crystal Structure of Dipeptide Epimerase From Enterococcus Faecalis V583 within 5.0Å range:
|
Magnesium binding site 2 out of 8 in 3jva
Go back to
Magnesium binding site 2 out
of 8 in the Crystal Structure of Dipeptide Epimerase From Enterococcus Faecalis V583
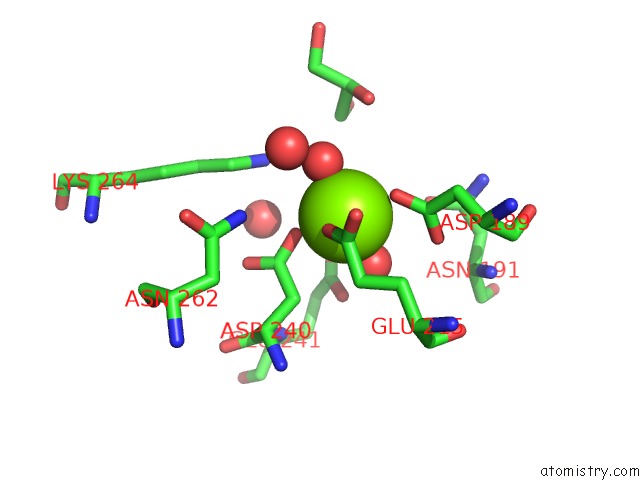
Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 2 of Crystal Structure of Dipeptide Epimerase From Enterococcus Faecalis V583 within 5.0Å range:
|
Magnesium binding site 3 out of 8 in 3jva
Go back to
Magnesium binding site 3 out
of 8 in the Crystal Structure of Dipeptide Epimerase From Enterococcus Faecalis V583

Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 3 of Crystal Structure of Dipeptide Epimerase From Enterococcus Faecalis V583 within 5.0Å range:
|
Magnesium binding site 4 out of 8 in 3jva
Go back to
Magnesium binding site 4 out
of 8 in the Crystal Structure of Dipeptide Epimerase From Enterococcus Faecalis V583
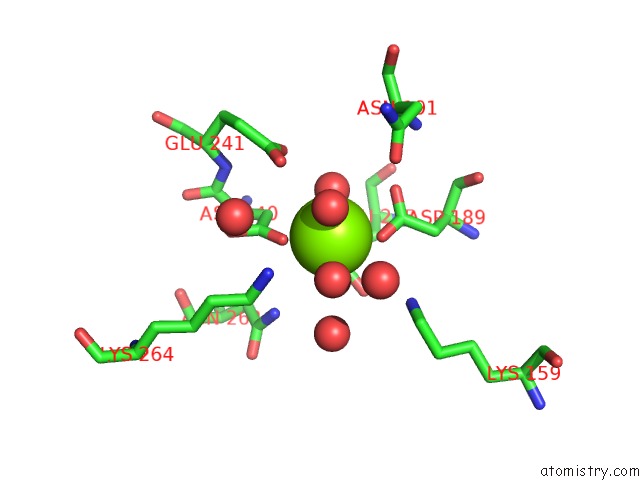
Mono view
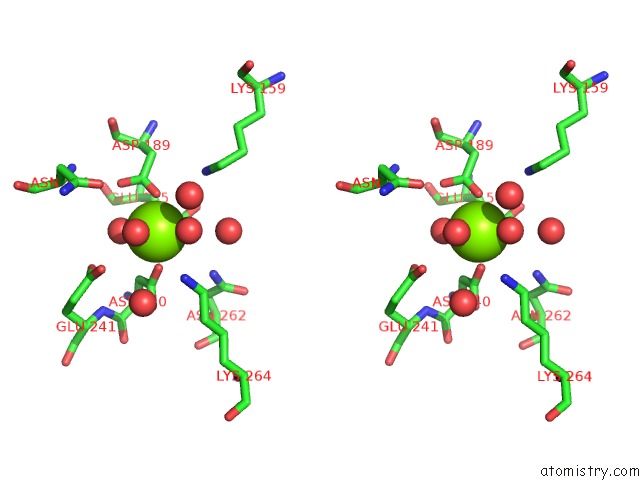
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 4 of Crystal Structure of Dipeptide Epimerase From Enterococcus Faecalis V583 within 5.0Å range:
|
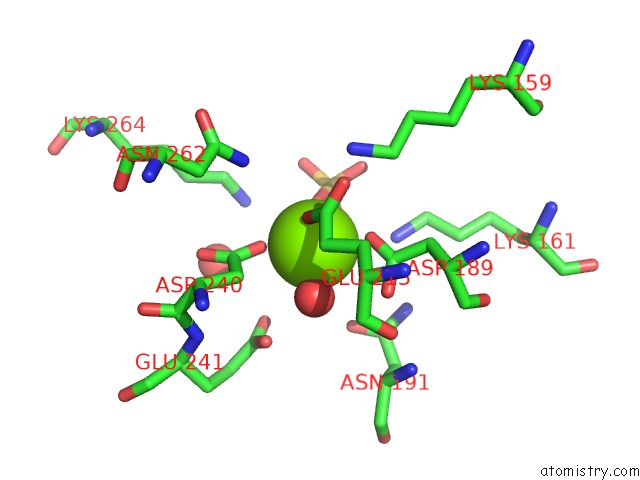
Magnesium binding site 5 out of 8 in 3jva
Go back to
Magnesium binding site 5 out
of 8 in the Crystal Structure of Dipeptide Epimerase From Enterococcus Faecalis V583
Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 5 of Crystal Structure of Dipeptide Epimerase From Enterococcus Faecalis V583 within 5.0Å range:
|
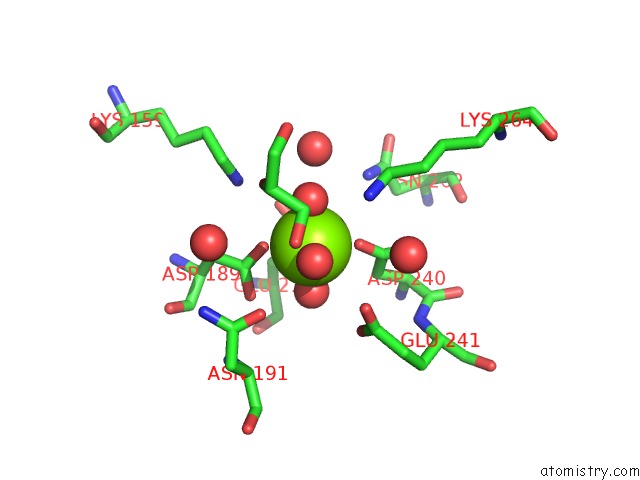
Magnesium binding site 6 out of 8 in 3jva
Go back to
Magnesium binding site 6 out
of 8 in the Crystal Structure of Dipeptide Epimerase From Enterococcus Faecalis V583
Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 6 of Crystal Structure of Dipeptide Epimerase From Enterococcus Faecalis V583 within 5.0Å range:
|
Magnesium binding site 7 out of 8 in 3jva
Go back to
Magnesium binding site 7 out
of 8 in the Crystal Structure of Dipeptide Epimerase From Enterococcus Faecalis V583

Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 7 of Crystal Structure of Dipeptide Epimerase From Enterococcus Faecalis V583 within 5.0Å range:
|
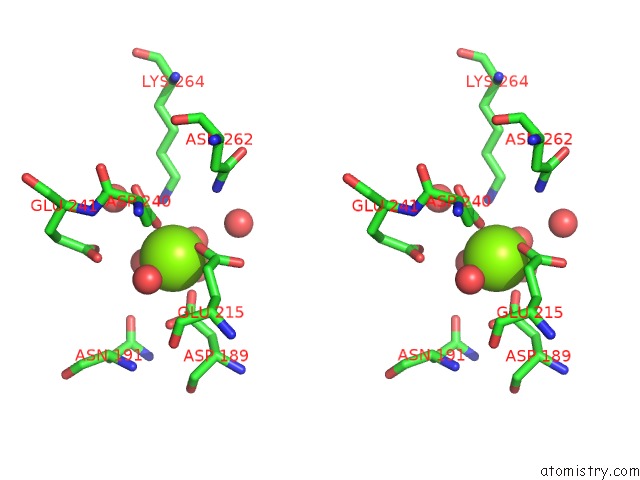
Magnesium binding site 8 out of 8 in 3jva
Go back to
Magnesium binding site 8 out
of 8 in the Crystal Structure of Dipeptide Epimerase From Enterococcus Faecalis V583

Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 8 of Crystal Structure of Dipeptide Epimerase From Enterococcus Faecalis V583 within 5.0Å range:
|
Reference:
T.Lukk,
A.Sakai,
C.Kalyanaraman,
S.D.Brown,
H.J.Imker,
L.Song,
A.A.Fedorov,
E.V.Fedorov,
R.Toro,
B.Hillerich,
R.Seidel,
Y.Patskovsky,
M.W.Vetting,
S.K.Nair,
P.C.Babbitt,
S.C.Almo,
J.A.Gerlt,
M.P.Jacobson.
Homology Models Guide Discovery of Diverse Enzyme Specificities Among Dipeptide Epimerases in the Enolase Superfamily. Proc.Natl.Acad.Sci.Usa V. 109 4122 2012.
ISSN: ISSN 0027-8424
PubMed: 22392983
DOI: 10.1073/PNAS.1112081109
Page generated: Sun Aug 10 23:21:09 2025
ISSN: ISSN 0027-8424
PubMed: 22392983
DOI: 10.1073/PNAS.1112081109
Last articles
Mg in 6WOAMg in 6WO9
Mg in 6WO8
Mg in 6WNR
Mg in 6WNQ
Mg in 6WO7
Mg in 6WO1
Mg in 6WMT
Mg in 6WMU
Mg in 6WMR