Magnesium »
PDB 5avz-5bjo »
5b4k »
Magnesium in PDB 5b4k: Crystal Structure of the Catalytic Domain of Human PDE10A Complexed with N-(4-((5-Methyl-5H-Pyrrolo[3,2-D]Pyrimidin-4-Yl)Oxy)Phenyl)-1H- Benzimidazol-2-Amine
Enzymatic activity of Crystal Structure of the Catalytic Domain of Human PDE10A Complexed with N-(4-((5-Methyl-5H-Pyrrolo[3,2-D]Pyrimidin-4-Yl)Oxy)Phenyl)-1H- Benzimidazol-2-Amine
All present enzymatic activity of Crystal Structure of the Catalytic Domain of Human PDE10A Complexed with N-(4-((5-Methyl-5H-Pyrrolo[3,2-D]Pyrimidin-4-Yl)Oxy)Phenyl)-1H- Benzimidazol-2-Amine:
3.1.4.17;
3.1.4.17;
Protein crystallography data
The structure of Crystal Structure of the Catalytic Domain of Human PDE10A Complexed with N-(4-((5-Methyl-5H-Pyrrolo[3,2-D]Pyrimidin-4-Yl)Oxy)Phenyl)-1H- Benzimidazol-2-Amine, PDB code: 5b4k
was solved by
H.Oki,
Y.Zama,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 80.83 / 2.90 |
| Space group | P 21 21 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 49.734, 81.683, 161.654, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 17.7 / 26.1 |
Other elements in 5b4k:
The structure of Crystal Structure of the Catalytic Domain of Human PDE10A Complexed with N-(4-((5-Methyl-5H-Pyrrolo[3,2-D]Pyrimidin-4-Yl)Oxy)Phenyl)-1H- Benzimidazol-2-Amine also contains other interesting chemical elements:
| Zinc | (Zn) | 2 atoms |
Magnesium Binding Sites:
The binding sites of Magnesium atom in the Crystal Structure of the Catalytic Domain of Human PDE10A Complexed with N-(4-((5-Methyl-5H-Pyrrolo[3,2-D]Pyrimidin-4-Yl)Oxy)Phenyl)-1H- Benzimidazol-2-Amine
(pdb code 5b4k). This binding sites where shown within
5.0 Angstroms radius around Magnesium atom.
In total 2 binding sites of Magnesium where determined in the Crystal Structure of the Catalytic Domain of Human PDE10A Complexed with N-(4-((5-Methyl-5H-Pyrrolo[3,2-D]Pyrimidin-4-Yl)Oxy)Phenyl)-1H- Benzimidazol-2-Amine, PDB code: 5b4k:
Jump to Magnesium binding site number: 1; 2;
In total 2 binding sites of Magnesium where determined in the Crystal Structure of the Catalytic Domain of Human PDE10A Complexed with N-(4-((5-Methyl-5H-Pyrrolo[3,2-D]Pyrimidin-4-Yl)Oxy)Phenyl)-1H- Benzimidazol-2-Amine, PDB code: 5b4k:
Jump to Magnesium binding site number: 1; 2;
Magnesium binding site 1 out of 2 in 5b4k
Go back to
Magnesium binding site 1 out
of 2 in the Crystal Structure of the Catalytic Domain of Human PDE10A Complexed with N-(4-((5-Methyl-5H-Pyrrolo[3,2-D]Pyrimidin-4-Yl)Oxy)Phenyl)-1H- Benzimidazol-2-Amine
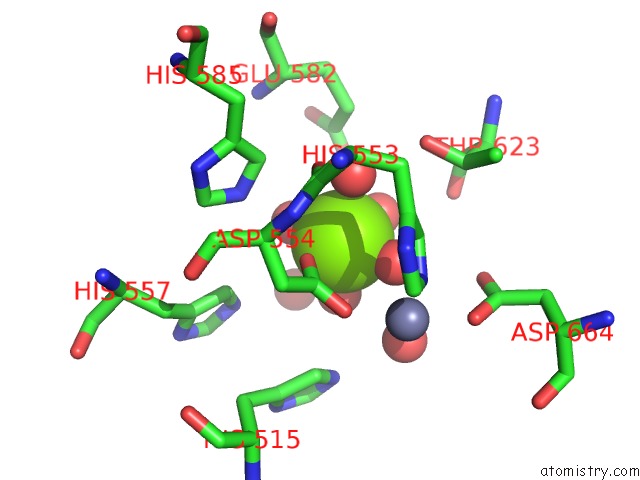
Mono view
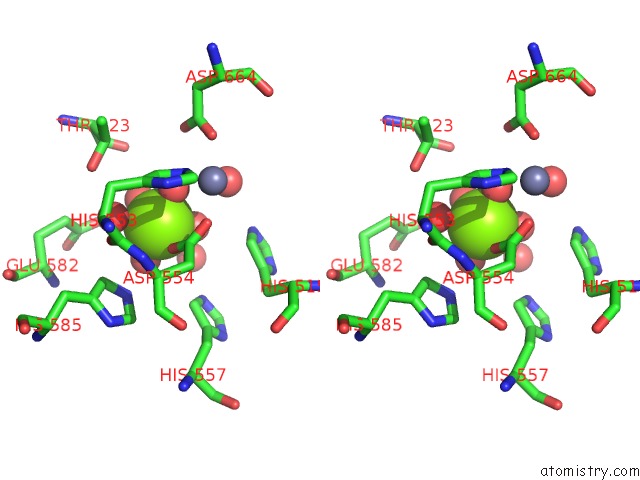
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 1 of Crystal Structure of the Catalytic Domain of Human PDE10A Complexed with N-(4-((5-Methyl-5H-Pyrrolo[3,2-D]Pyrimidin-4-Yl)Oxy)Phenyl)-1H- Benzimidazol-2-Amine within 5.0Å range:
|
Magnesium binding site 2 out of 2 in 5b4k
Go back to
Magnesium binding site 2 out
of 2 in the Crystal Structure of the Catalytic Domain of Human PDE10A Complexed with N-(4-((5-Methyl-5H-Pyrrolo[3,2-D]Pyrimidin-4-Yl)Oxy)Phenyl)-1H- Benzimidazol-2-Amine
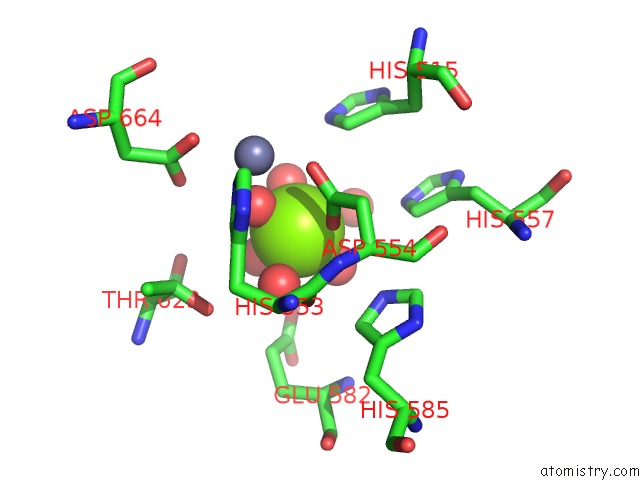
Mono view
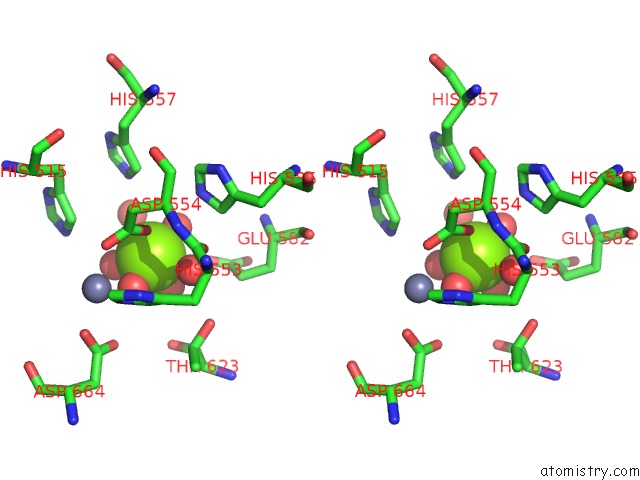
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 2 of Crystal Structure of the Catalytic Domain of Human PDE10A Complexed with N-(4-((5-Methyl-5H-Pyrrolo[3,2-D]Pyrimidin-4-Yl)Oxy)Phenyl)-1H- Benzimidazol-2-Amine within 5.0Å range:
|
Reference:
M.Yoshikawa,
T.Hitaka,
T.Hasui,
M.Fushimi,
J.Kunitomo,
H.Kokubo,
H.Oki,
K.Nakashima,
T.Taniguchi.
Design and Synthesis of Potent and Selective Pyridazin-4(1H)-One-Based PDE10A Inhibitors Interacting with TYR683 in the PDE10A Selectivity Pocket Bioorg.Med.Chem. V. 24 3447 2016.
ISSN: ESSN 1464-3391
PubMed: 27301679
DOI: 10.1016/J.BMC.2016.05.049
Page generated: Tue Aug 12 05:33:04 2025
ISSN: ESSN 1464-3391
PubMed: 27301679
DOI: 10.1016/J.BMC.2016.05.049
Last articles
Mg in 5L5XMg in 5L5V
Mg in 5L5U
Mg in 5L5T
Mg in 5L5S
Mg in 5L5R
Mg in 5L5P
Mg in 5L5Q
Mg in 5L5O
Mg in 5L5J