Magnesium »
PDB 5cr7-5d2j »
5d1o »
Magnesium in PDB 5d1o: Archaeal Atp-Dependent Rna Ligase - Form 1
Protein crystallography data
The structure of Archaeal Atp-Dependent Rna Ligase - Form 1, PDB code: 5d1o
was solved by
K.S.Murakami,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 27.71 / 2.65 |
| Space group | P 1 21 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 50.250, 114.870, 91.290, 90.00, 104.42, 90.00 |
| R / Rfree (%) | 20.6 / 26.5 |
Magnesium Binding Sites:
The binding sites of Magnesium atom in the Archaeal Atp-Dependent Rna Ligase - Form 1
(pdb code 5d1o). This binding sites where shown within
5.0 Angstroms radius around Magnesium atom.
In total only one binding site of Magnesium was determined in the Archaeal Atp-Dependent Rna Ligase - Form 1, PDB code: 5d1o:
In total only one binding site of Magnesium was determined in the Archaeal Atp-Dependent Rna Ligase - Form 1, PDB code: 5d1o:
Magnesium binding site 1 out of 1 in 5d1o
Go back to
Magnesium binding site 1 out
of 1 in the Archaeal Atp-Dependent Rna Ligase - Form 1
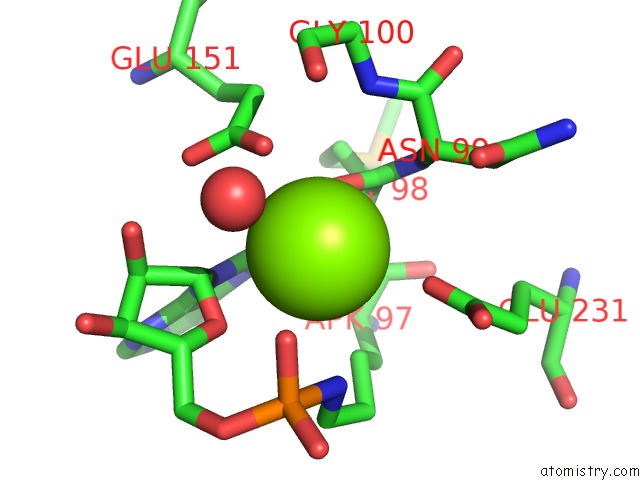
Mono view
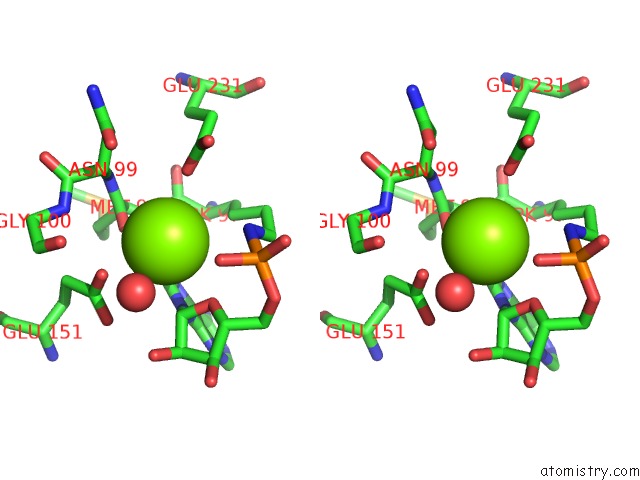
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 1 of Archaeal Atp-Dependent Rna Ligase - Form 1 within 5.0Å range:
|
Reference:
H.Gu,
S.Yoshinari,
R.Ghosh,
A.V.Ignatochkina,
P.D.Gollnick,
K.S.Murakami,
C.K.Ho.
Structural and Mutational Analysis of Archaeal Atp-Dependent Rna Ligase Identifies Amino Acids Required For Rna Binding and Catalysis. Nucleic Acids Res. V. 44 2337 2016.
ISSN: ESSN 1362-4962
PubMed: 26896806
DOI: 10.1093/NAR/GKW094
Page generated: Sun Sep 29 02:27:08 2024
ISSN: ESSN 1362-4962
PubMed: 26896806
DOI: 10.1093/NAR/GKW094
Last articles
K in 9DITK in 9DJV
K in 9DIG
K in 9DID
K in 9DIC
K in 9DI8
K in 9DIB
K in 9DE5
K in 9D5W
K in 9D19