Magnesium »
PDB 5grf-5h2f »
5gz9 »
Magnesium in PDB 5gz9: Crystal Structure of Catalytic Domain of Protein O-Mannosyl Kinase in Complexes with Amp-Pnp, Magnesium Ions and Glycopeptide
Protein crystallography data
The structure of Crystal Structure of Catalytic Domain of Protein O-Mannosyl Kinase in Complexes with Amp-Pnp, Magnesium Ions and Glycopeptide, PDB code: 5gz9
was solved by
M.Nagae,
Y.Yamaguchi,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 40.19 / 2.40 |
| Space group | P 31 2 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 81.320, 81.320, 146.838, 90.00, 90.00, 120.00 |
| R / Rfree (%) | 23.2 / 26.8 |
Magnesium Binding Sites:
The binding sites of Magnesium atom in the Crystal Structure of Catalytic Domain of Protein O-Mannosyl Kinase in Complexes with Amp-Pnp, Magnesium Ions and Glycopeptide
(pdb code 5gz9). This binding sites where shown within
5.0 Angstroms radius around Magnesium atom.
In total 2 binding sites of Magnesium where determined in the Crystal Structure of Catalytic Domain of Protein O-Mannosyl Kinase in Complexes with Amp-Pnp, Magnesium Ions and Glycopeptide, PDB code: 5gz9:
Jump to Magnesium binding site number: 1; 2;
In total 2 binding sites of Magnesium where determined in the Crystal Structure of Catalytic Domain of Protein O-Mannosyl Kinase in Complexes with Amp-Pnp, Magnesium Ions and Glycopeptide, PDB code: 5gz9:
Jump to Magnesium binding site number: 1; 2;
Magnesium binding site 1 out of 2 in 5gz9
Go back to
Magnesium binding site 1 out
of 2 in the Crystal Structure of Catalytic Domain of Protein O-Mannosyl Kinase in Complexes with Amp-Pnp, Magnesium Ions and Glycopeptide
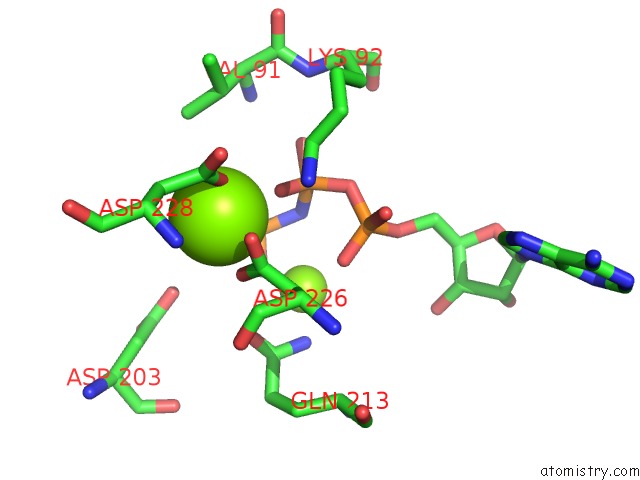
Mono view
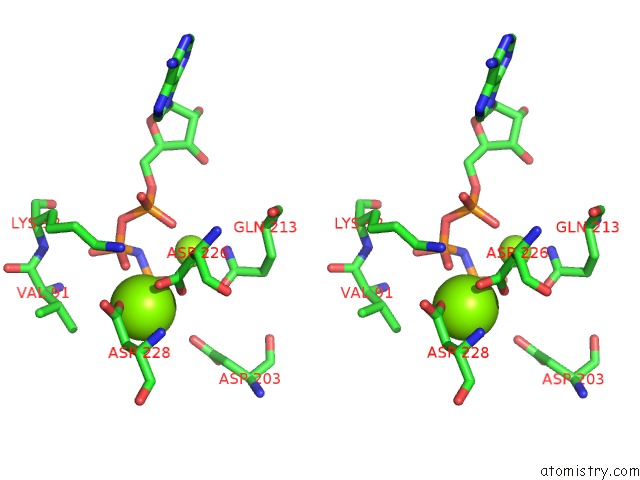
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 1 of Crystal Structure of Catalytic Domain of Protein O-Mannosyl Kinase in Complexes with Amp-Pnp, Magnesium Ions and Glycopeptide within 5.0Å range:
|
Magnesium binding site 2 out of 2 in 5gz9
Go back to
Magnesium binding site 2 out
of 2 in the Crystal Structure of Catalytic Domain of Protein O-Mannosyl Kinase in Complexes with Amp-Pnp, Magnesium Ions and Glycopeptide
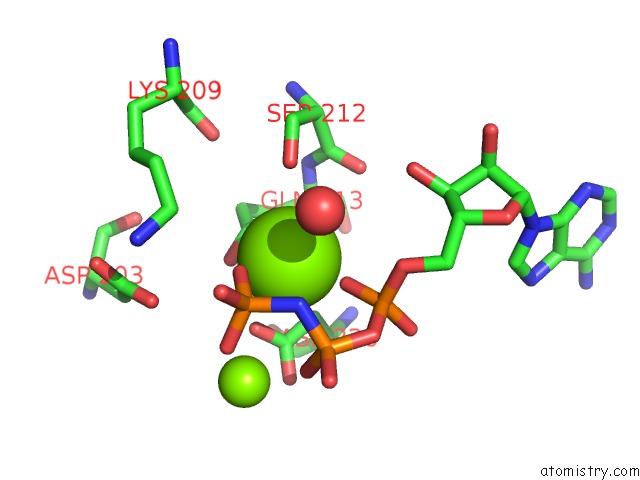
Mono view
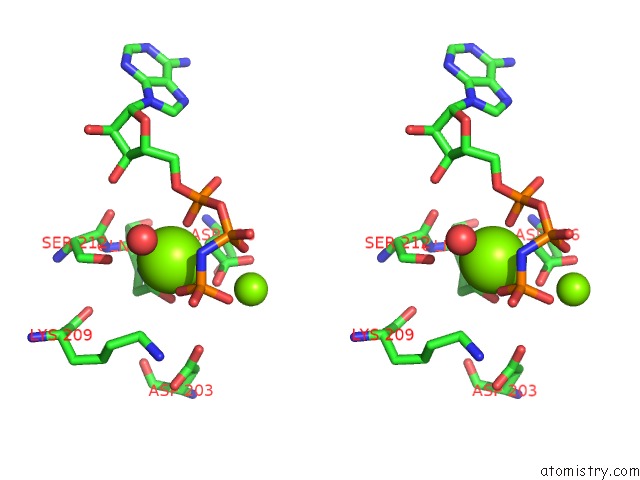
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 2 of Crystal Structure of Catalytic Domain of Protein O-Mannosyl Kinase in Complexes with Amp-Pnp, Magnesium Ions and Glycopeptide within 5.0Å range:
|
Reference:
M.Nagae,
S.K.Mishra,
M.Neyazaki,
R.Oi,
A.Ikeda,
N.Matsugaki,
S.Akashi,
H.Manya,
M.Mizuno,
H.Yagi,
K.Kato,
T.Senda,
T.Endo,
T.Nogi,
Y.Yamaguchi.
3D Structural Analysis of Protein O-Mannosyl Kinase, Pomk, A Causative Gene Product of Dystroglycanopathy. Genes Cells V. 22 348 2017.
ISSN: ESSN 1365-2443
PubMed: 28251761
DOI: 10.1111/GTC.12480
Page generated: Sun Sep 29 15:31:23 2024
ISSN: ESSN 1365-2443
PubMed: 28251761
DOI: 10.1111/GTC.12480
Last articles
K in 8JAHK in 8IAX
K in 8IPP
K in 8IBN
K in 8IAU
K in 8I03
K in 8IAT
K in 8I02
K in 8HZF
K in 8HZM