Magnesium »
PDB 5u7d-5uhd »
5uah »
Magnesium in PDB 5uah: Escherichia Coli Rna Polymerase and Rifampin Complex, Rpob D516V Mutant
Enzymatic activity of Escherichia Coli Rna Polymerase and Rifampin Complex, Rpob D516V Mutant
All present enzymatic activity of Escherichia Coli Rna Polymerase and Rifampin Complex, Rpob D516V Mutant:
2.7.7.6;
2.7.7.6;
Protein crystallography data
The structure of Escherichia Coli Rna Polymerase and Rifampin Complex, Rpob D516V Mutant, PDB code: 5uah
was solved by
V.Molodtsov,
N.T.Scharf,
M.A.Stefan,
G.A.Garcia,
K.S.Murakami,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 29.74 / 4.10 |
| Space group | P 21 21 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 187.814, 206.668, 310.286, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 24.3 / 28.7 |
Other elements in 5uah:
The structure of Escherichia Coli Rna Polymerase and Rifampin Complex, Rpob D516V Mutant also contains other interesting chemical elements:
| Zinc | (Zn) | 4 atoms |
Magnesium Binding Sites:
The binding sites of Magnesium atom in the Escherichia Coli Rna Polymerase and Rifampin Complex, Rpob D516V Mutant
(pdb code 5uah). This binding sites where shown within
5.0 Angstroms radius around Magnesium atom.
In total 4 binding sites of Magnesium where determined in the Escherichia Coli Rna Polymerase and Rifampin Complex, Rpob D516V Mutant, PDB code: 5uah:
Jump to Magnesium binding site number: 1; 2; 3; 4;
In total 4 binding sites of Magnesium where determined in the Escherichia Coli Rna Polymerase and Rifampin Complex, Rpob D516V Mutant, PDB code: 5uah:
Jump to Magnesium binding site number: 1; 2; 3; 4;
Magnesium binding site 1 out of 4 in 5uah
Go back to
Magnesium binding site 1 out
of 4 in the Escherichia Coli Rna Polymerase and Rifampin Complex, Rpob D516V Mutant
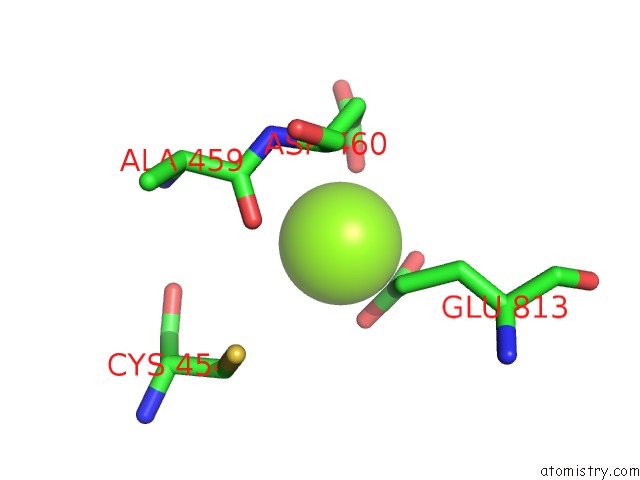
Mono view
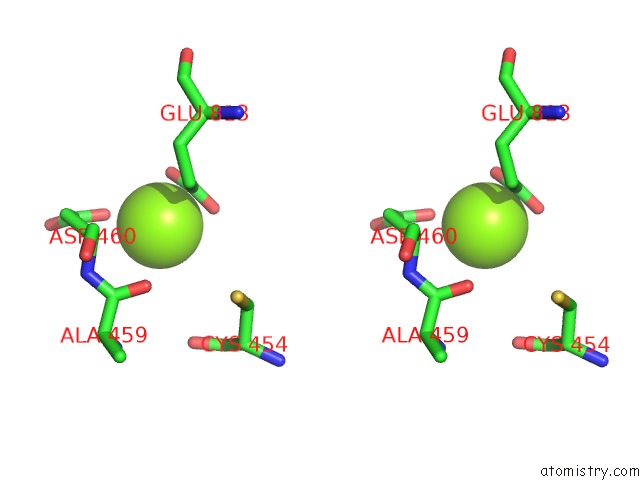
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 1 of Escherichia Coli Rna Polymerase and Rifampin Complex, Rpob D516V Mutant within 5.0Å range:
|
Magnesium binding site 2 out of 4 in 5uah
Go back to
Magnesium binding site 2 out
of 4 in the Escherichia Coli Rna Polymerase and Rifampin Complex, Rpob D516V Mutant
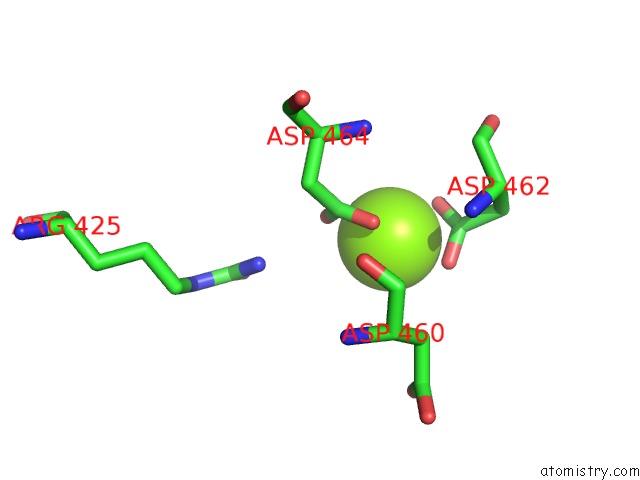
Mono view
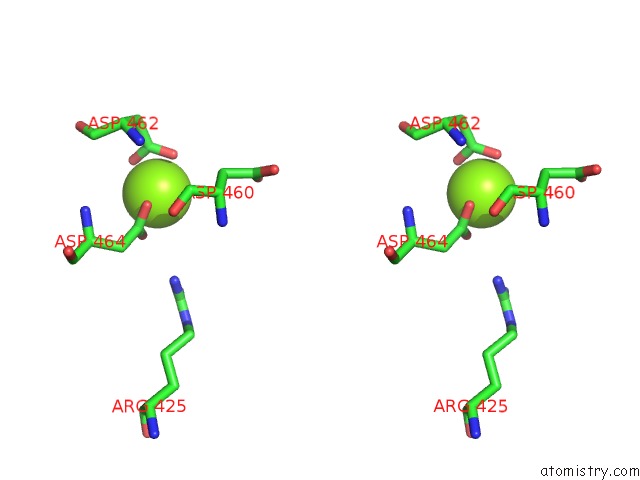
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 2 of Escherichia Coli Rna Polymerase and Rifampin Complex, Rpob D516V Mutant within 5.0Å range:
|
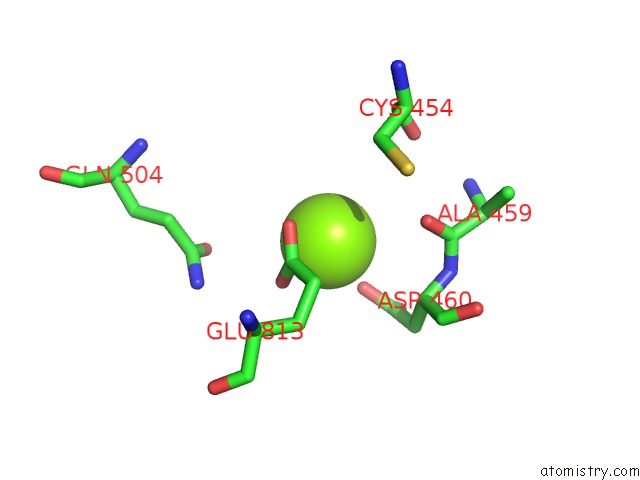
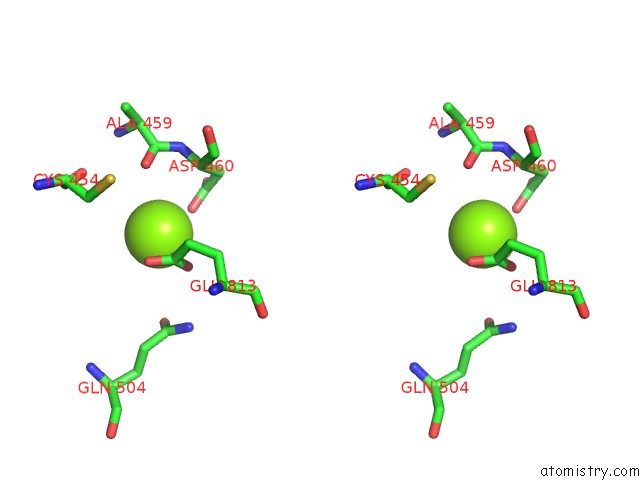
Magnesium binding site 3 out of 4 in 5uah
Go back to
Magnesium binding site 3 out
of 4 in the Escherichia Coli Rna Polymerase and Rifampin Complex, Rpob D516V Mutant
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 3 of Escherichia Coli Rna Polymerase and Rifampin Complex, Rpob D516V Mutant within 5.0Å range:
|
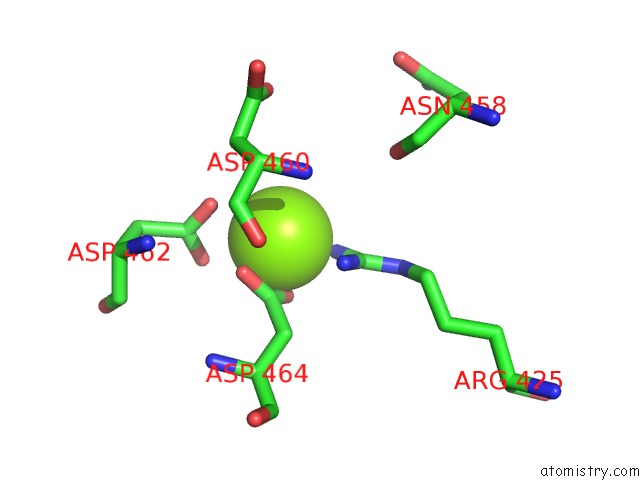
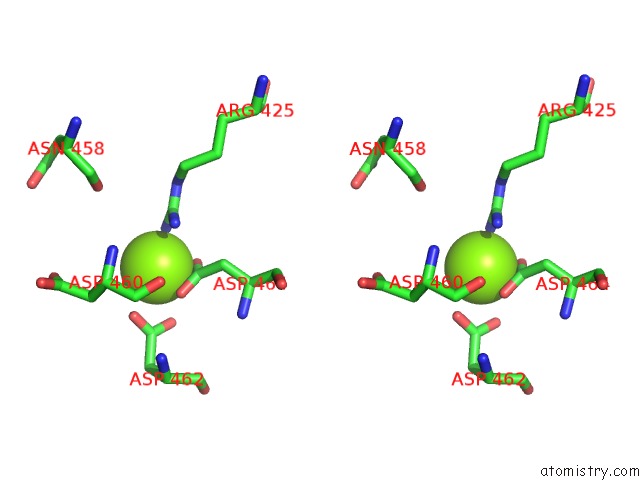
Magnesium binding site 4 out of 4 in 5uah
Go back to
Magnesium binding site 4 out
of 4 in the Escherichia Coli Rna Polymerase and Rifampin Complex, Rpob D516V Mutant
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 4 of Escherichia Coli Rna Polymerase and Rifampin Complex, Rpob D516V Mutant within 5.0Å range:
|
Reference:
V.Molodtsov,
N.T.Scharf,
M.A.Stefan,
G.A.Garcia,
K.S.Murakami.
Structural Basis For Rifamycin Resistance of Bacterial Rna Polymerase By the Three Most Clinically Important Rpob Mutations Found in Mycobacterium Tuberculosis. Mol. Microbiol. V. 103 1034 2017.
ISSN: ESSN 1365-2958
PubMed: 28009073
DOI: 10.1111/MMI.13606
Page generated: Tue Aug 12 20:46:57 2025
ISSN: ESSN 1365-2958
PubMed: 28009073
DOI: 10.1111/MMI.13606
Last articles
Mg in 6BR1Mg in 6BR7
Mg in 6BQZ
Mg in 6BNQ
Mg in 6BQF
Mg in 6BOJ
Mg in 6BQ2
Mg in 6BNP
Mg in 6BNO
Mg in 6BNJ