Magnesium »
PDB 5xlh-5xut »
5xp8 »
Magnesium in PDB 5xp8: Crystal Structure of T. Thermophilus Argonaute Protein Complexed with A Bulge 4A5 on the Guide Strand
Protein crystallography data
The structure of Crystal Structure of T. Thermophilus Argonaute Protein Complexed with A Bulge 4A5 on the Guide Strand, PDB code: 5xp8
was solved by
G.Sheng,
T.Gogakos,
J.Wang,
H.Zhao,
A.Serganov,
S.Juranek,
T.Tuschl,
J.D.Patel,
Y.Wang,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 45.20 / 3.10 |
| Space group | P 41 3 2 |
| Cell size a, b, c (Å), α, β, γ (°) | 202.140, 202.140, 202.140, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 21.4 / 27.6 |
Magnesium Binding Sites:
The binding sites of Magnesium atom in the Crystal Structure of T. Thermophilus Argonaute Protein Complexed with A Bulge 4A5 on the Guide Strand
(pdb code 5xp8). This binding sites where shown within
5.0 Angstroms radius around Magnesium atom.
In total 3 binding sites of Magnesium where determined in the Crystal Structure of T. Thermophilus Argonaute Protein Complexed with A Bulge 4A5 on the Guide Strand, PDB code: 5xp8:
Jump to Magnesium binding site number: 1; 2; 3;
In total 3 binding sites of Magnesium where determined in the Crystal Structure of T. Thermophilus Argonaute Protein Complexed with A Bulge 4A5 on the Guide Strand, PDB code: 5xp8:
Jump to Magnesium binding site number: 1; 2; 3;
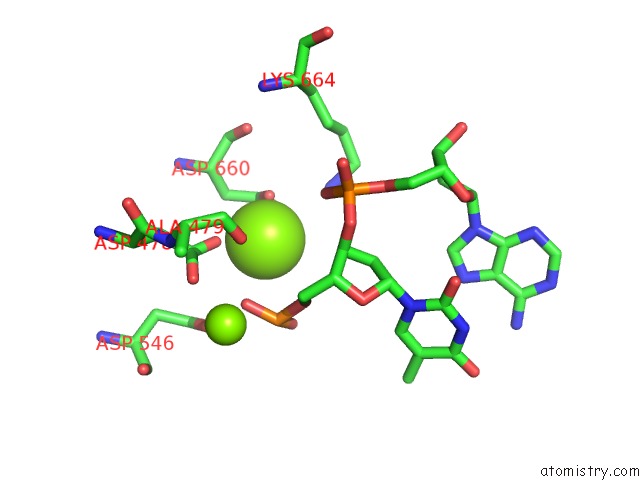
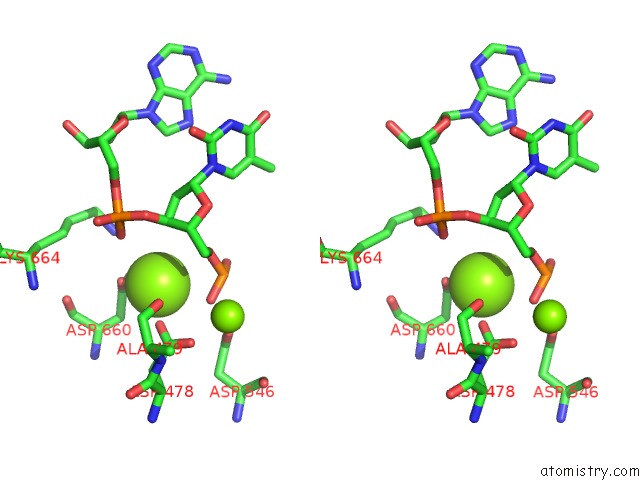
Magnesium binding site 1 out of 3 in 5xp8
Go back to
Magnesium binding site 1 out
of 3 in the Crystal Structure of T. Thermophilus Argonaute Protein Complexed with A Bulge 4A5 on the Guide Strand
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 1 of Crystal Structure of T. Thermophilus Argonaute Protein Complexed with A Bulge 4A5 on the Guide Strand within 5.0Å range:
|
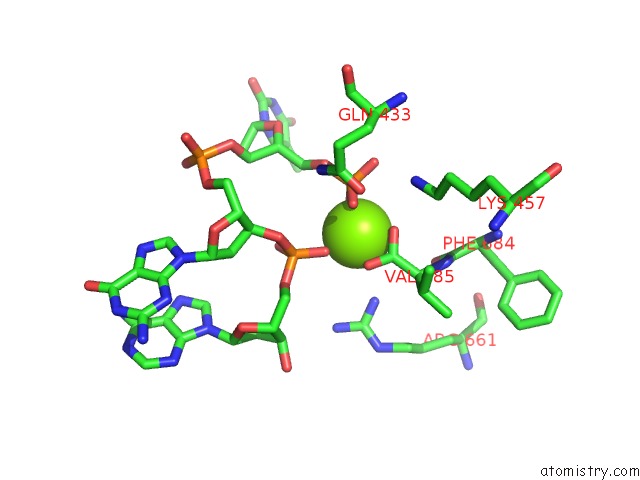
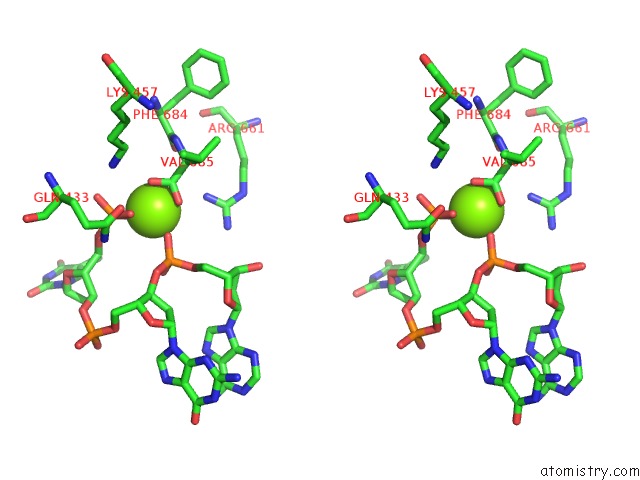
Magnesium binding site 2 out of 3 in 5xp8
Go back to
Magnesium binding site 2 out
of 3 in the Crystal Structure of T. Thermophilus Argonaute Protein Complexed with A Bulge 4A5 on the Guide Strand
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 2 of Crystal Structure of T. Thermophilus Argonaute Protein Complexed with A Bulge 4A5 on the Guide Strand within 5.0Å range:
|
Magnesium binding site 3 out of 3 in 5xp8
Go back to
Magnesium binding site 3 out
of 3 in the Crystal Structure of T. Thermophilus Argonaute Protein Complexed with A Bulge 4A5 on the Guide Strand
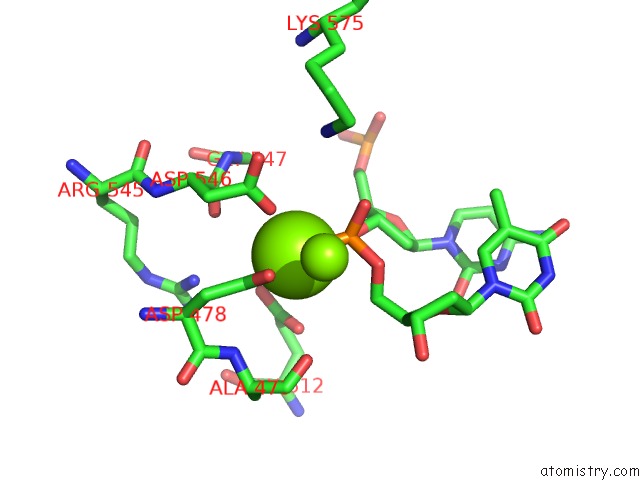
Mono view
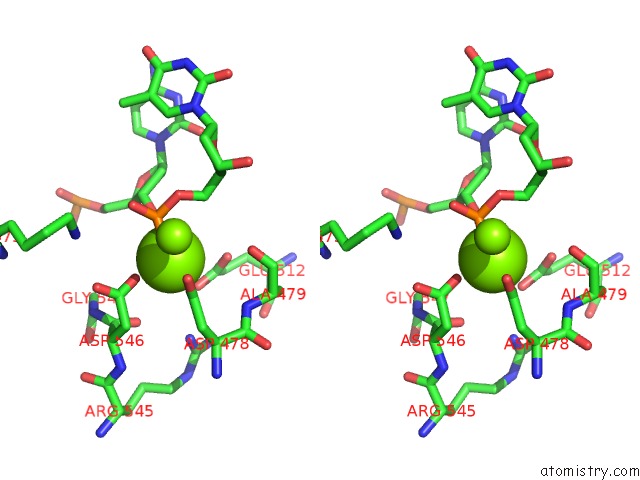
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 3 of Crystal Structure of T. Thermophilus Argonaute Protein Complexed with A Bulge 4A5 on the Guide Strand within 5.0Å range:
|
Reference:
G.Sheng,
T.Gogakos,
J.Wang,
H.Zhao,
A.Serganov,
S.Juranek,
T.Tuschl,
D.J.Patel,
Y.Wang.
Structure/Cleavage-Based Insights Into Helical Perturbations at Bulge Sites Within T. Thermophilus Argonaute Silencing Complexes. Nucleic Acids Res. V. 45 9149 2017.
ISSN: ESSN 1362-4962
PubMed: 28911094
DOI: 10.1093/NAR/GKX547
Page generated: Tue Aug 12 23:57:10 2025
ISSN: ESSN 1362-4962
PubMed: 28911094
DOI: 10.1093/NAR/GKX547
Last articles
Mg in 6YHSMg in 6YEZ
Mg in 6YBW
Mg in 6YKW
Mg in 6YKV
Mg in 6YKU
Mg in 6YKT
Mg in 6YKS
Mg in 6YKQ
Mg in 6YKO