Magnesium »
PDB 5zlf-6a1m »
5zri »
Magnesium in PDB 5zri: M. Smegmatis Antimutator Protein MUTT2 in Complex with 5M-Dcmp
Protein crystallography data
The structure of M. Smegmatis Antimutator Protein MUTT2 in Complex with 5M-Dcmp, PDB code: 5zri
was solved by
A.Singh,
S.M.Arif,
P.B.Sang,
U.Varshney,
M.Vijayan,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 29.42 / 1.58 |
| Space group | P 21 21 2 |
| Cell size a, b, c (Å), α, β, γ (°) | 65.520, 58.840, 31.350, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 17.7 / 21.1 |
Magnesium Binding Sites:
The binding sites of Magnesium atom in the M. Smegmatis Antimutator Protein MUTT2 in Complex with 5M-Dcmp
(pdb code 5zri). This binding sites where shown within
5.0 Angstroms radius around Magnesium atom.
In total only one binding site of Magnesium was determined in the M. Smegmatis Antimutator Protein MUTT2 in Complex with 5M-Dcmp, PDB code: 5zri:
In total only one binding site of Magnesium was determined in the M. Smegmatis Antimutator Protein MUTT2 in Complex with 5M-Dcmp, PDB code: 5zri:
Magnesium binding site 1 out of 1 in 5zri
Go back to
Magnesium binding site 1 out
of 1 in the M. Smegmatis Antimutator Protein MUTT2 in Complex with 5M-Dcmp
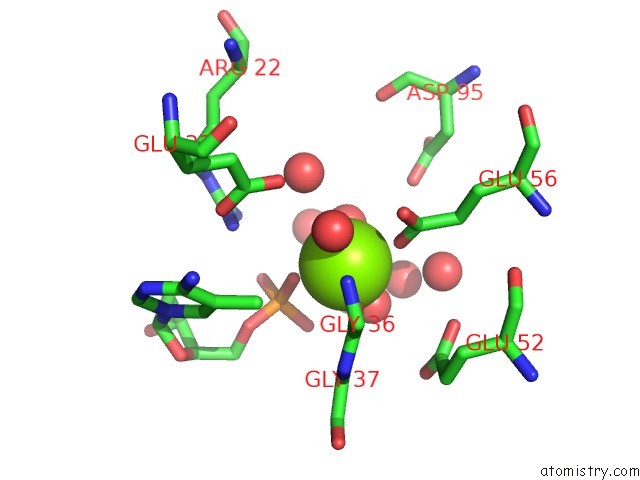
Mono view
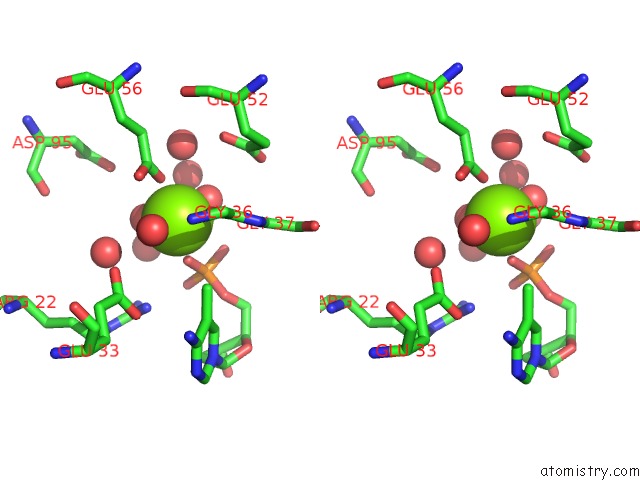
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 1 of M. Smegmatis Antimutator Protein MUTT2 in Complex with 5M-Dcmp within 5.0Å range:
|
Reference:
A.Singh,
S.Mohammad Arif,
P.Biak Sang,
U.Varshney,
M.Vijayan.
Structural Insights Into the Specificity and Catalytic Mechanism of Mycobacterial Nucleotide Pool Sanitizing Enzyme MUTT2. J.Struct.Biol. V. 204 449 2018.
ISSN: ESSN 1095-8657
PubMed: 30312643
DOI: 10.1016/J.JSB.2018.10.002
Page generated: Wed Aug 13 01:53:05 2025
ISSN: ESSN 1095-8657
PubMed: 30312643
DOI: 10.1016/J.JSB.2018.10.002
Last articles
Mg in 6QPHMg in 6QV0
Mg in 6QUZ
Mg in 6QUY
Mg in 6QUX
Mg in 6QUW
Mg in 6QUV
Mg in 6QUU
Mg in 6QUS
Mg in 6QTN