Magnesium »
PDB 6f6u-6fch »
6f95 »
Magnesium in PDB 6f95: Alfa From B. Subtilis Plasmid PLS32 Filament Structure at 3.4 A
Magnesium Binding Sites:
The binding sites of Magnesium atom in the Alfa From B. Subtilis Plasmid PLS32 Filament Structure at 3.4 A
(pdb code 6f95). This binding sites where shown within
5.0 Angstroms radius around Magnesium atom.
In total 5 binding sites of Magnesium where determined in the Alfa From B. Subtilis Plasmid PLS32 Filament Structure at 3.4 A, PDB code: 6f95:
Jump to Magnesium binding site number: 1; 2; 3; 4; 5;
In total 5 binding sites of Magnesium where determined in the Alfa From B. Subtilis Plasmid PLS32 Filament Structure at 3.4 A, PDB code: 6f95:
Jump to Magnesium binding site number: 1; 2; 3; 4; 5;
Magnesium binding site 1 out of 5 in 6f95
Go back to
Magnesium binding site 1 out
of 5 in the Alfa From B. Subtilis Plasmid PLS32 Filament Structure at 3.4 A
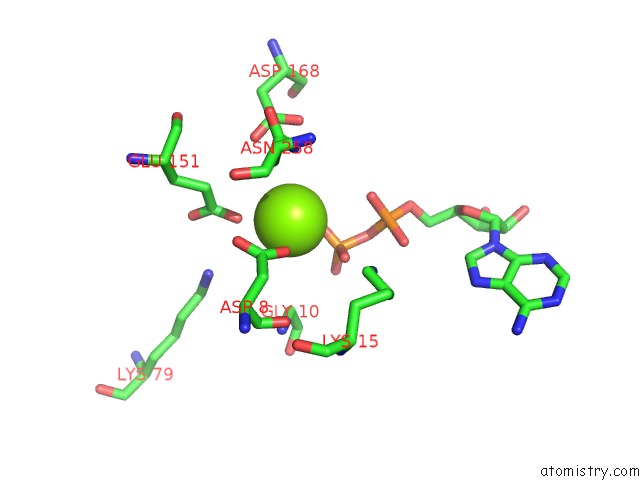
Mono view
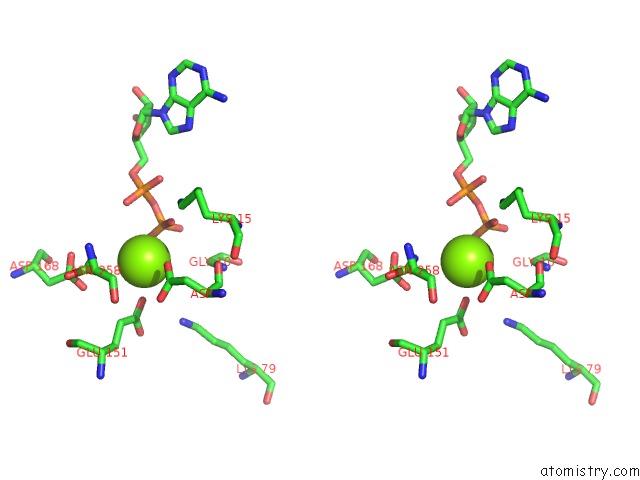
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 1 of Alfa From B. Subtilis Plasmid PLS32 Filament Structure at 3.4 A within 5.0Å range:
|
Magnesium binding site 2 out of 5 in 6f95
Go back to
Magnesium binding site 2 out
of 5 in the Alfa From B. Subtilis Plasmid PLS32 Filament Structure at 3.4 A
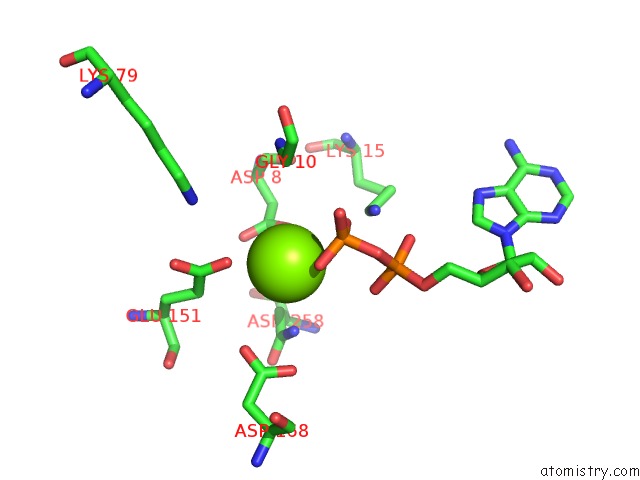
Mono view
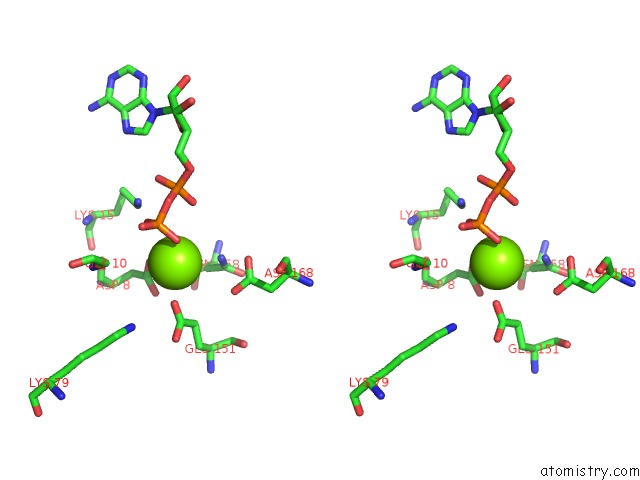
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 2 of Alfa From B. Subtilis Plasmid PLS32 Filament Structure at 3.4 A within 5.0Å range:
|
Magnesium binding site 3 out of 5 in 6f95
Go back to
Magnesium binding site 3 out
of 5 in the Alfa From B. Subtilis Plasmid PLS32 Filament Structure at 3.4 A
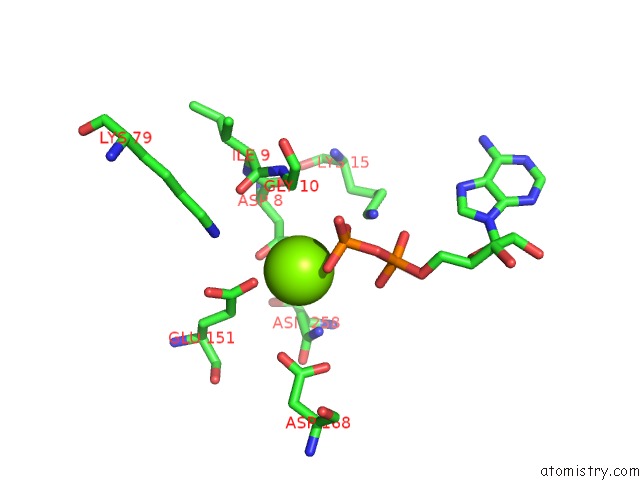
Mono view
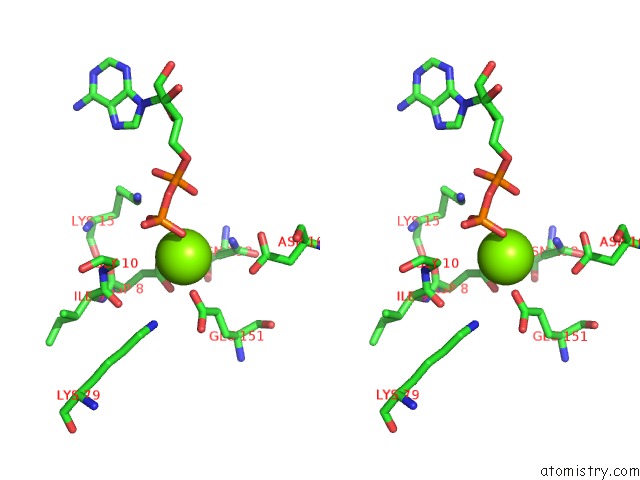
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 3 of Alfa From B. Subtilis Plasmid PLS32 Filament Structure at 3.4 A within 5.0Å range:
|
Magnesium binding site 4 out of 5 in 6f95
Go back to
Magnesium binding site 4 out
of 5 in the Alfa From B. Subtilis Plasmid PLS32 Filament Structure at 3.4 A
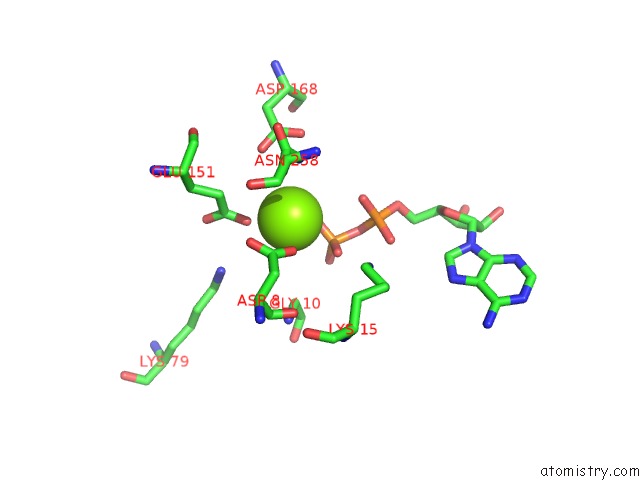
Mono view
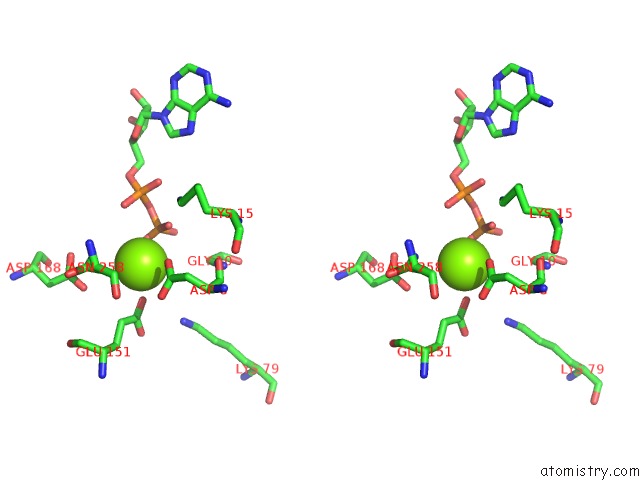
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 4 of Alfa From B. Subtilis Plasmid PLS32 Filament Structure at 3.4 A within 5.0Å range:
|
Magnesium binding site 5 out of 5 in 6f95
Go back to
Magnesium binding site 5 out
of 5 in the Alfa From B. Subtilis Plasmid PLS32 Filament Structure at 3.4 A
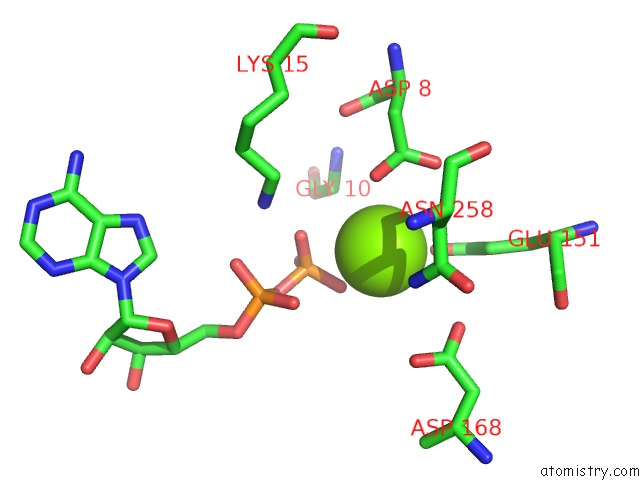
Mono view
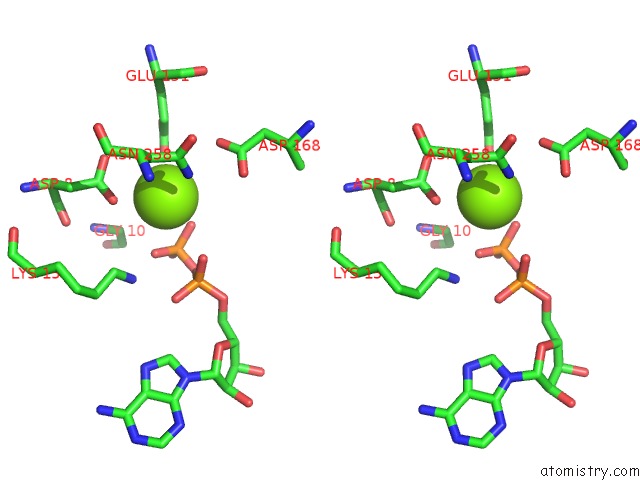
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 5 of Alfa From B. Subtilis Plasmid PLS32 Filament Structure at 3.4 A within 5.0Å range:
|
Reference:
A.Szewczak-Harris,
J.Lowe.
Cryo-Em Reconstruction of Alfa Frombacillus Subtilisreveals the Structure of A Simplified Actin-Like Filament at 3.4- Angstrom Resolution. Proc. Natl. Acad. Sci. V. 115 3458 2018U.S.A..
ISSN: ESSN 1091-6490
PubMed: 29440489
DOI: 10.1073/PNAS.1716424115
Page generated: Tue Oct 1 00:14:09 2024
ISSN: ESSN 1091-6490
PubMed: 29440489
DOI: 10.1073/PNAS.1716424115
Last articles
K in 9CBRK in 9CBP
K in 9C91
K in 9C0H
K in 9C0J
K in 9C09
K in 9C0E
K in 9C0I
K in 9BZ1
K in 9BZC