Magnesium »
PDB 6gov-6gvc »
6gs2 »
Magnesium in PDB 6gs2: Crystal Structure of the Gatd/Murt Enzyme Complex From Staphylococcus Aureus
Protein crystallography data
The structure of Crystal Structure of the Gatd/Murt Enzyme Complex From Staphylococcus Aureus, PDB code: 6gs2
was solved by
L.M.Muckenfuss,
E.R.Noeldeke,
V.Niemann,
G.Zocher,
T.Stehle,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 49.06 / 2.04 |
| Space group | P 21 21 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 107.100, 110.370, 116.360, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 17.5 / 21.6 |
Other elements in 6gs2:
The structure of Crystal Structure of the Gatd/Murt Enzyme Complex From Staphylococcus Aureus also contains other interesting chemical elements:
| Zinc | (Zn) | 2 atoms |
Magnesium Binding Sites:
The binding sites of Magnesium atom in the Crystal Structure of the Gatd/Murt Enzyme Complex From Staphylococcus Aureus
(pdb code 6gs2). This binding sites where shown within
5.0 Angstroms radius around Magnesium atom.
In total only one binding site of Magnesium was determined in the Crystal Structure of the Gatd/Murt Enzyme Complex From Staphylococcus Aureus, PDB code: 6gs2:
In total only one binding site of Magnesium was determined in the Crystal Structure of the Gatd/Murt Enzyme Complex From Staphylococcus Aureus, PDB code: 6gs2:
Magnesium binding site 1 out of 1 in 6gs2
Go back to
Magnesium binding site 1 out
of 1 in the Crystal Structure of the Gatd/Murt Enzyme Complex From Staphylococcus Aureus
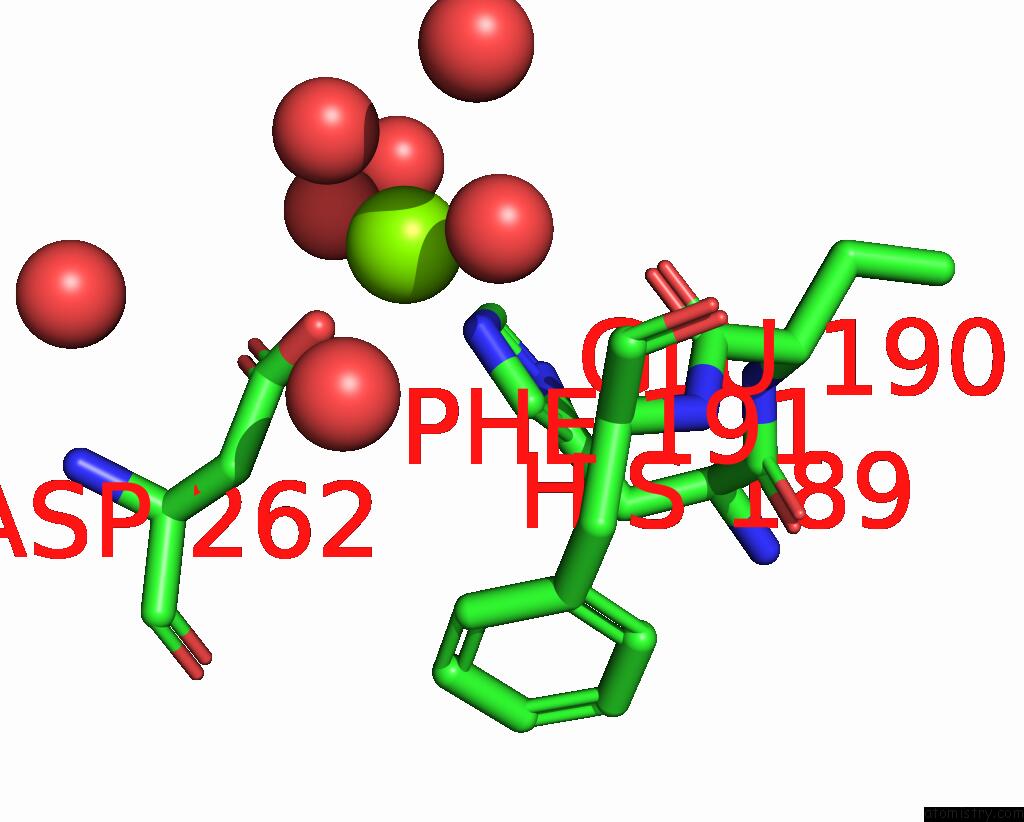
Mono view
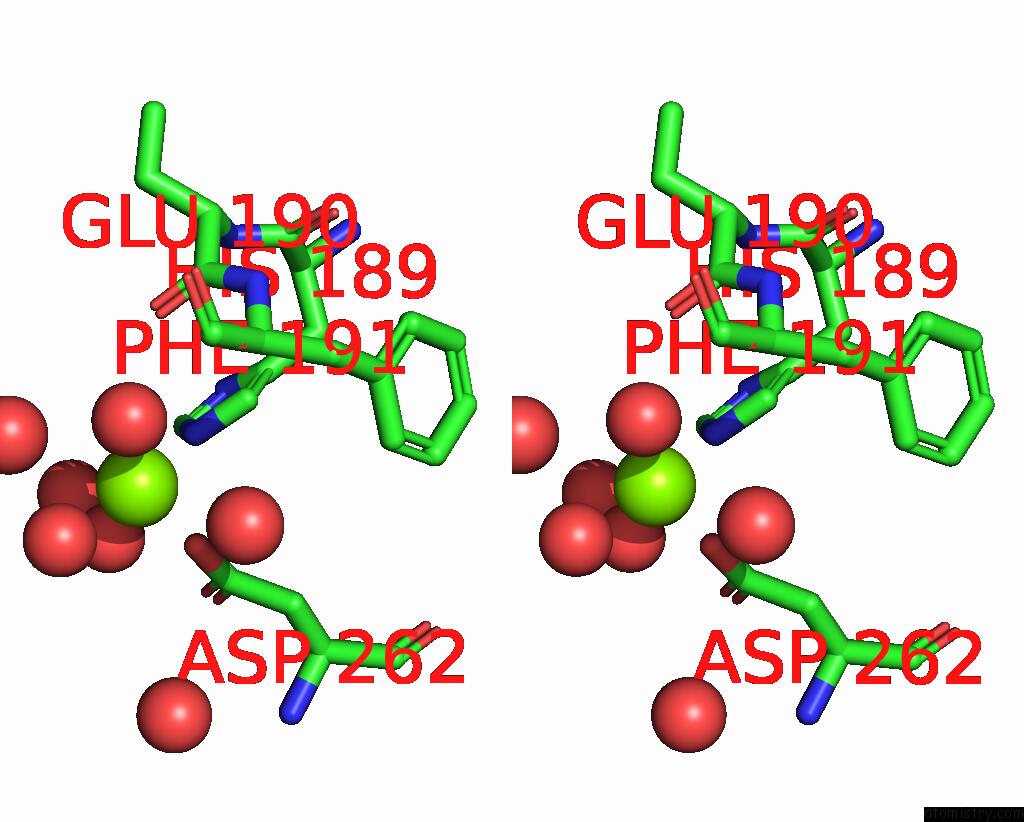
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 1 of Crystal Structure of the Gatd/Murt Enzyme Complex From Staphylococcus Aureus within 5.0Å range:
|
Reference:
E.R.Noldeke,
L.M.Muckenfuss,
V.Niemann,
A.Muller,
E.Stork,
G.Zocher,
T.Schneider,
T.Stehle.
Structural Basis of Cell Wall Peptidoglycan Amidation By the Gatd/Murt Complex of Staphylococcus Aureus. Sci Rep V. 8 12953 2018.
ISSN: ESSN 2045-2322
PubMed: 30154570
DOI: 10.1038/S41598-018-31098-X
Page generated: Wed Aug 13 06:44:50 2025
ISSN: ESSN 2045-2322
PubMed: 30154570
DOI: 10.1038/S41598-018-31098-X
Last articles
Mg in 9JMAMg in 9JKP
Mg in 9JJW
Mg in 9JJV
Mg in 9JI2
Mg in 9JFJ
Mg in 9J89
Mg in 9JC1
Mg in 9JAO
Mg in 9JA1