Magnesium »
PDB 6jkm-6jws »
6jp4 »
Magnesium in PDB 6jp4: Crystal Structure of the Catalytic Domain of A Multi-Domain Alginate Lyase DP0100 From Thermophilic Bacterium Defluviitalea Phaphyphila
Protein crystallography data
The structure of Crystal Structure of the Catalytic Domain of A Multi-Domain Alginate Lyase DP0100 From Thermophilic Bacterium Defluviitalea Phaphyphila, PDB code: 6jp4
was solved by
S.Q.Ji,
S.R.Dix,
A.Aziz,
S.E.Sedelnikova,
F.L.Li,
D.W.Rice,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 98.90 / 2.07 |
| Space group | C 2 2 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 261.324, 394.812, 112.187, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 22.2 / 24 |
Other elements in 6jp4:
The structure of Crystal Structure of the Catalytic Domain of A Multi-Domain Alginate Lyase DP0100 From Thermophilic Bacterium Defluviitalea Phaphyphila also contains other interesting chemical elements:
| Manganese | (Mn) | 3 atoms |
| Arsenic | (As) | 1 atom |
| Calcium | (Ca) | 6 atoms |
Magnesium Binding Sites:
The binding sites of Magnesium atom in the Crystal Structure of the Catalytic Domain of A Multi-Domain Alginate Lyase DP0100 From Thermophilic Bacterium Defluviitalea Phaphyphila
(pdb code 6jp4). This binding sites where shown within
5.0 Angstroms radius around Magnesium atom.
In total 3 binding sites of Magnesium where determined in the Crystal Structure of the Catalytic Domain of A Multi-Domain Alginate Lyase DP0100 From Thermophilic Bacterium Defluviitalea Phaphyphila, PDB code: 6jp4:
Jump to Magnesium binding site number: 1; 2; 3;
In total 3 binding sites of Magnesium where determined in the Crystal Structure of the Catalytic Domain of A Multi-Domain Alginate Lyase DP0100 From Thermophilic Bacterium Defluviitalea Phaphyphila, PDB code: 6jp4:
Jump to Magnesium binding site number: 1; 2; 3;
Magnesium binding site 1 out of 3 in 6jp4
Go back to
Magnesium binding site 1 out
of 3 in the Crystal Structure of the Catalytic Domain of A Multi-Domain Alginate Lyase DP0100 From Thermophilic Bacterium Defluviitalea Phaphyphila
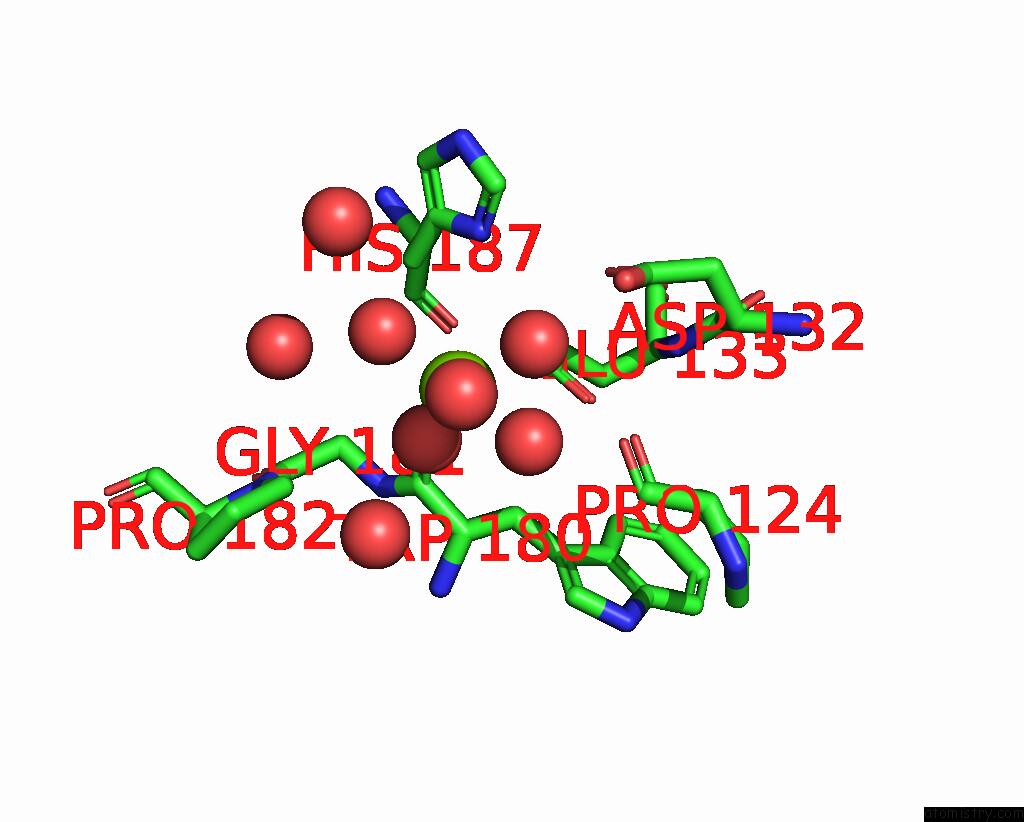
Mono view
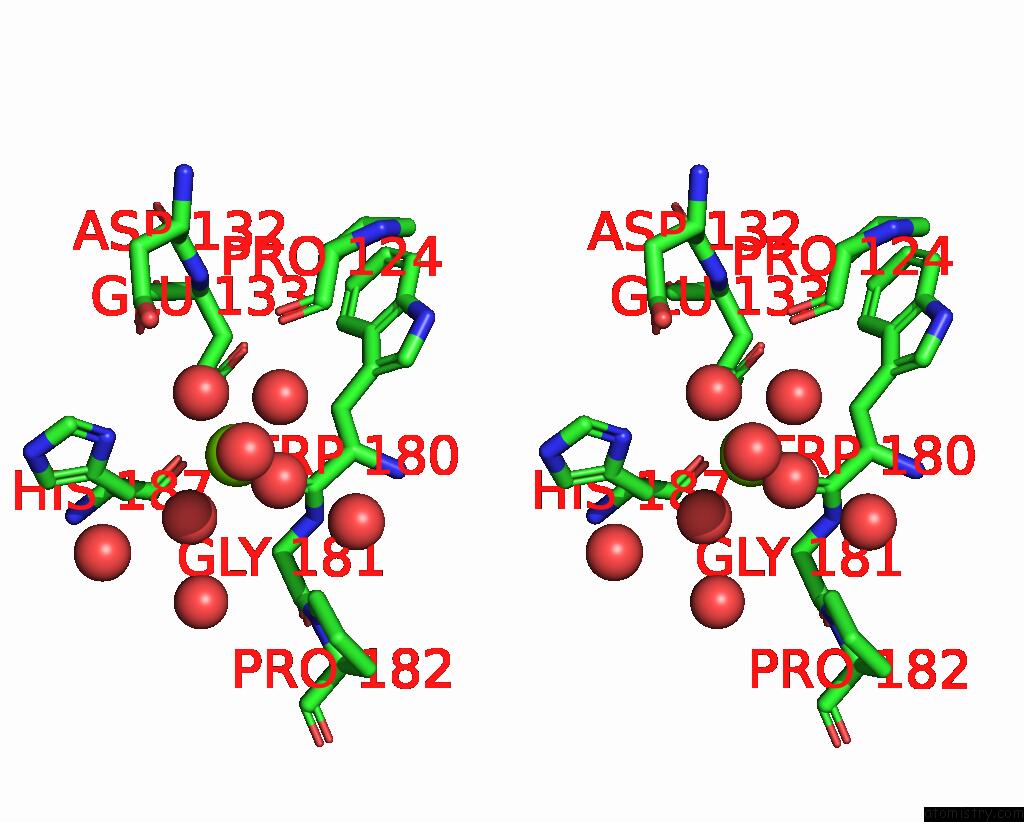
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 1 of Crystal Structure of the Catalytic Domain of A Multi-Domain Alginate Lyase DP0100 From Thermophilic Bacterium Defluviitalea Phaphyphila within 5.0Å range:
|
Magnesium binding site 2 out of 3 in 6jp4
Go back to
Magnesium binding site 2 out
of 3 in the Crystal Structure of the Catalytic Domain of A Multi-Domain Alginate Lyase DP0100 From Thermophilic Bacterium Defluviitalea Phaphyphila
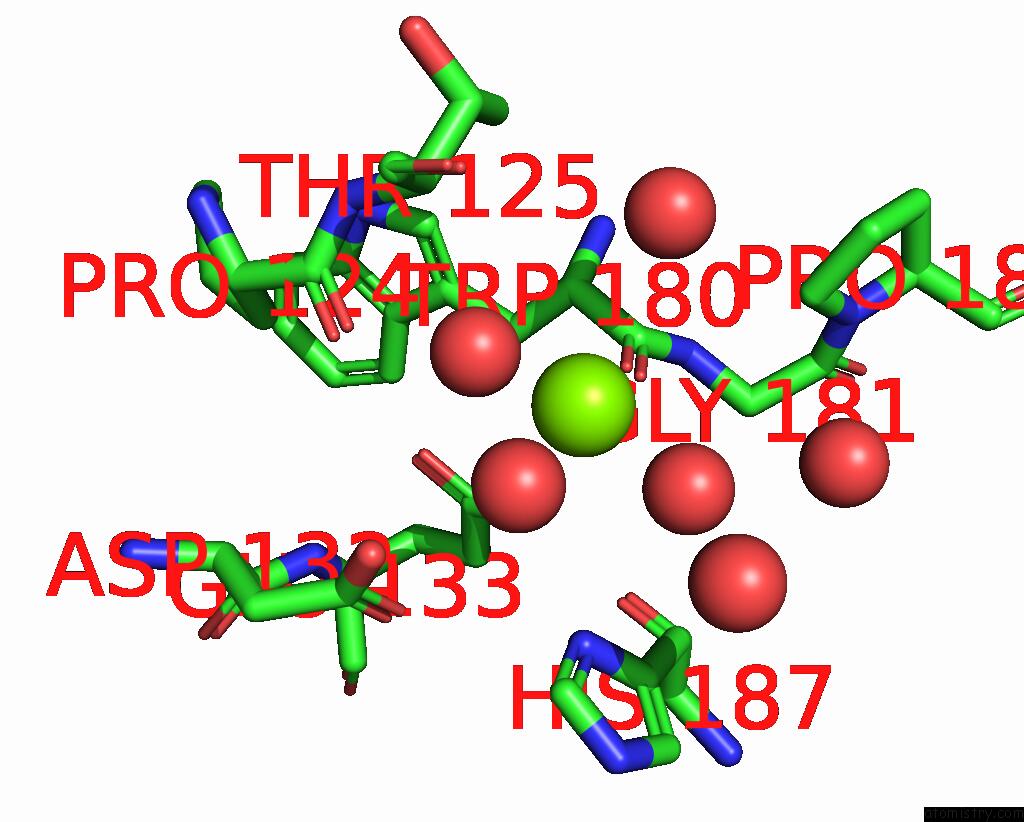
Mono view
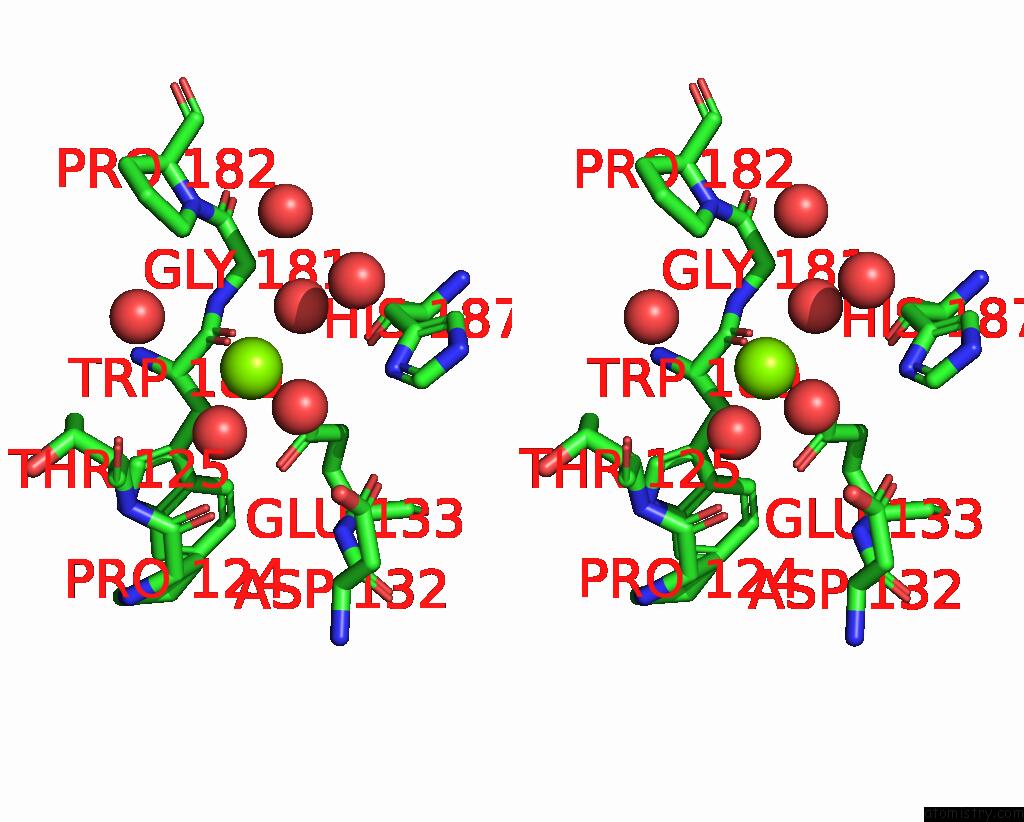
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 2 of Crystal Structure of the Catalytic Domain of A Multi-Domain Alginate Lyase DP0100 From Thermophilic Bacterium Defluviitalea Phaphyphila within 5.0Å range:
|
Magnesium binding site 3 out of 3 in 6jp4
Go back to
Magnesium binding site 3 out
of 3 in the Crystal Structure of the Catalytic Domain of A Multi-Domain Alginate Lyase DP0100 From Thermophilic Bacterium Defluviitalea Phaphyphila
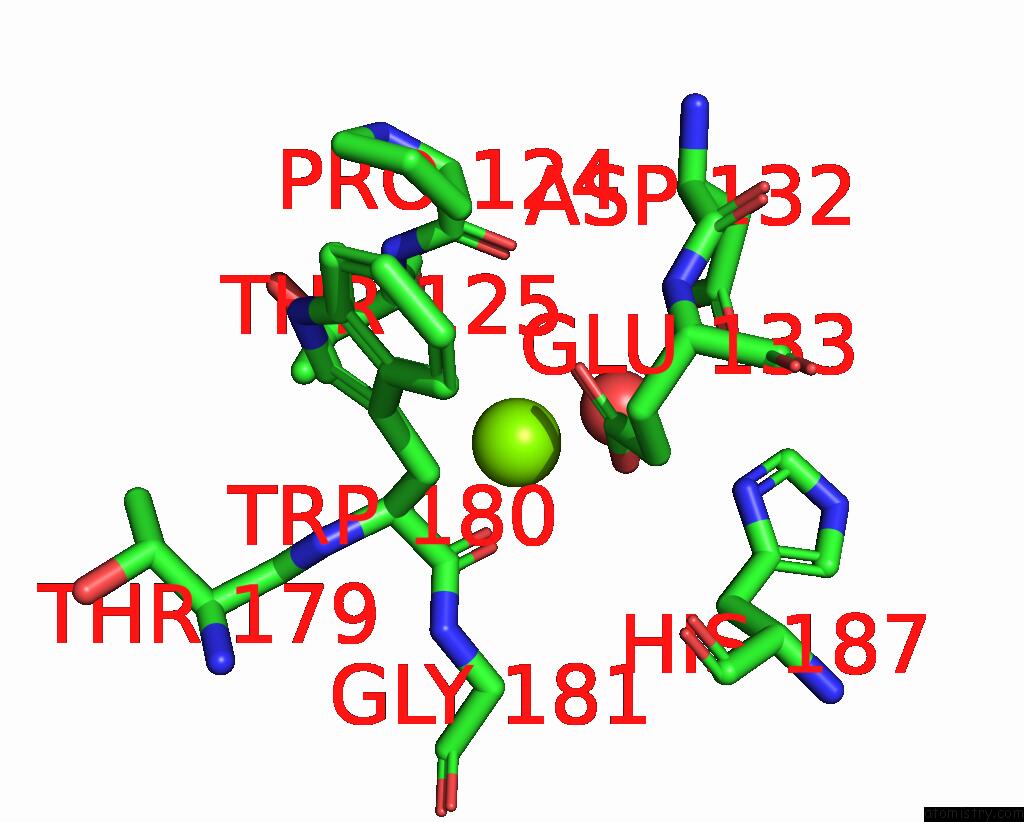
Mono view
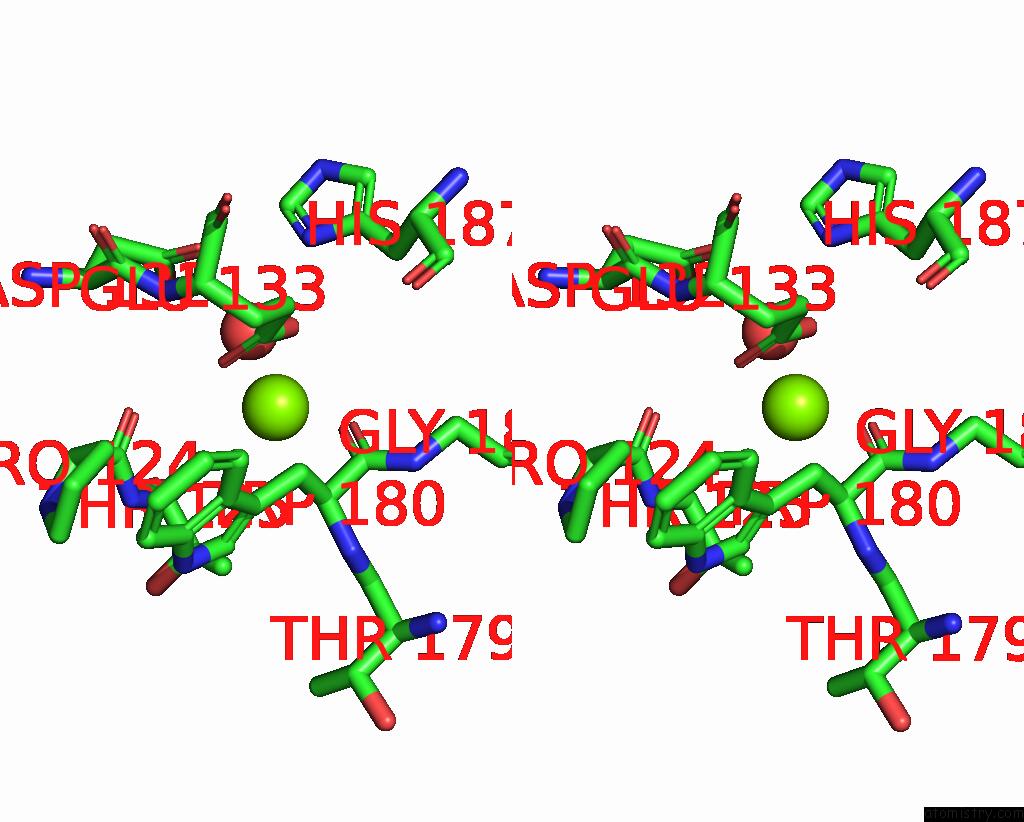
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 3 of Crystal Structure of the Catalytic Domain of A Multi-Domain Alginate Lyase DP0100 From Thermophilic Bacterium Defluviitalea Phaphyphila within 5.0Å range:
|
Reference:
S.Ji,
S.R.Dix,
A.A.Aziz,
S.E.Sedelnikova,
P.J.Baker,
J.B.Rafferty,
P.A.Bullough,
S.B.Tzokov,
J.Agirre,
F.L.Li,
D.W.Rice.
The Molecular Basis of Endolytic Activity of A Multidomain Alginate Lyase From Defluviitalea Phaphyphila, A Representative of A New Lyase Family, Plxx. J.Biol.Chem. 2019.
ISSN: ESSN 1083-351X
PubMed: 31624143
DOI: 10.1074/JBC.RA119.010716
Page generated: Tue Oct 1 05:44:08 2024
ISSN: ESSN 1083-351X
PubMed: 31624143
DOI: 10.1074/JBC.RA119.010716
Last articles
K in 2UXCK in 2UUC
K in 2VGB
K in 2VCG
K in 2VDD
K in 2V5X
K in 2V5W
K in 2UXD
K in 2UYY
K in 2UUB