Magnesium »
PDB 6l5l-6ll1 »
6lcr »
Magnesium in PDB 6lcr: Cryo-Em Structure of DNF1 From Chaetomium Thermophilum in the E1-Atp State
Enzymatic activity of Cryo-Em Structure of DNF1 From Chaetomium Thermophilum in the E1-Atp State
All present enzymatic activity of Cryo-Em Structure of DNF1 From Chaetomium Thermophilum in the E1-Atp State:
7.6.2.1;
7.6.2.1;
Magnesium Binding Sites:
The binding sites of Magnesium atom in the Cryo-Em Structure of DNF1 From Chaetomium Thermophilum in the E1-Atp State
(pdb code 6lcr). This binding sites where shown within
5.0 Angstroms radius around Magnesium atom.
In total only one binding site of Magnesium was determined in the Cryo-Em Structure of DNF1 From Chaetomium Thermophilum in the E1-Atp State, PDB code: 6lcr:
In total only one binding site of Magnesium was determined in the Cryo-Em Structure of DNF1 From Chaetomium Thermophilum in the E1-Atp State, PDB code: 6lcr:
Magnesium binding site 1 out of 1 in 6lcr
Go back to
Magnesium binding site 1 out
of 1 in the Cryo-Em Structure of DNF1 From Chaetomium Thermophilum in the E1-Atp State
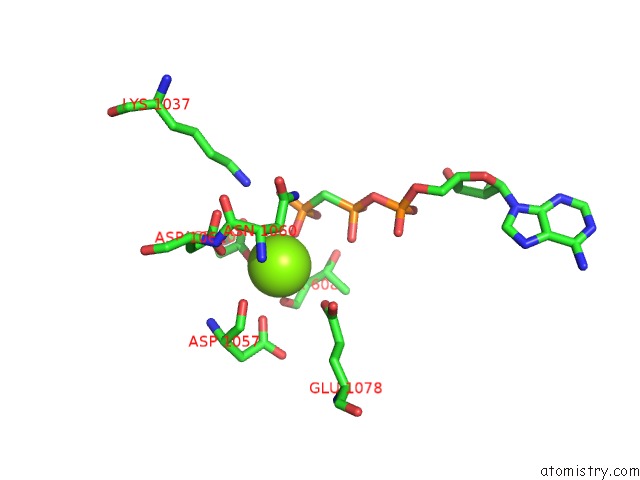
Mono view
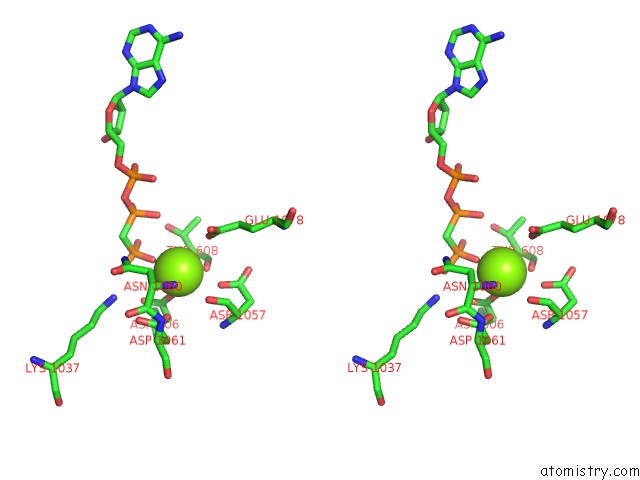
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 1 of Cryo-Em Structure of DNF1 From Chaetomium Thermophilum in the E1-Atp State within 5.0Å range:
|
Reference:
Y.He,
J.Xu,
X.Wu,
L.Li.
Structures of A P4-Atpase Lipid Flippase in Lipid Bilayers. Protein Cell 2020.
ISSN: ESSN 1674-8018
PubMed: 32303992
DOI: 10.1007/S13238-020-00712-Y
Page generated: Tue Oct 1 10:23:36 2024
ISSN: ESSN 1674-8018
PubMed: 32303992
DOI: 10.1007/S13238-020-00712-Y
Last articles
K in 4XTJK in 4XS4
K in 4XS1
K in 4XK0
K in 4XRD
K in 4XRA
K in 4XQK
K in 4XG1
K in 4XOU
K in 4XDL