Magnesium »
PDB 6mm5-6mxg »
6mo5 »
Magnesium in PDB 6mo5: Co-Crystal Structure of P. Aeruginosa Lpxc-50228 Complex
Enzymatic activity of Co-Crystal Structure of P. Aeruginosa Lpxc-50228 Complex
All present enzymatic activity of Co-Crystal Structure of P. Aeruginosa Lpxc-50228 Complex:
3.5.1.108;
3.5.1.108;
Protein crystallography data
The structure of Co-Crystal Structure of P. Aeruginosa Lpxc-50228 Complex, PDB code: 6mo5
was solved by
A.J.Stein,
M.C.Holt,
Z.Assar,
F.Cohen,
L.Andrews,
R.Cirz,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 33.24 / 1.85 |
| Space group | P 1 21 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 36.101, 66.486, 63.565, 90.00, 90.59, 90.00 |
| R / Rfree (%) | 17.7 / 21.9 |
Magnesium Binding Sites:
The binding sites of Magnesium atom in the Co-Crystal Structure of P. Aeruginosa Lpxc-50228 Complex
(pdb code 6mo5). This binding sites where shown within
5.0 Angstroms radius around Magnesium atom.
In total only one binding site of Magnesium was determined in the Co-Crystal Structure of P. Aeruginosa Lpxc-50228 Complex, PDB code: 6mo5:
In total only one binding site of Magnesium was determined in the Co-Crystal Structure of P. Aeruginosa Lpxc-50228 Complex, PDB code: 6mo5:
Magnesium binding site 1 out of 1 in 6mo5
Go back to
Magnesium binding site 1 out
of 1 in the Co-Crystal Structure of P. Aeruginosa Lpxc-50228 Complex
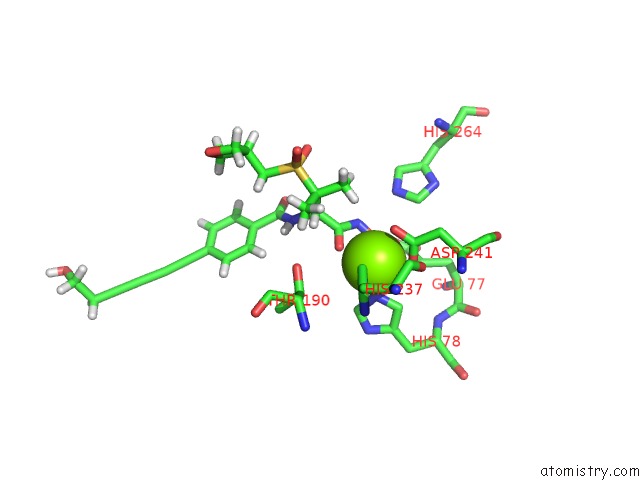
Mono view
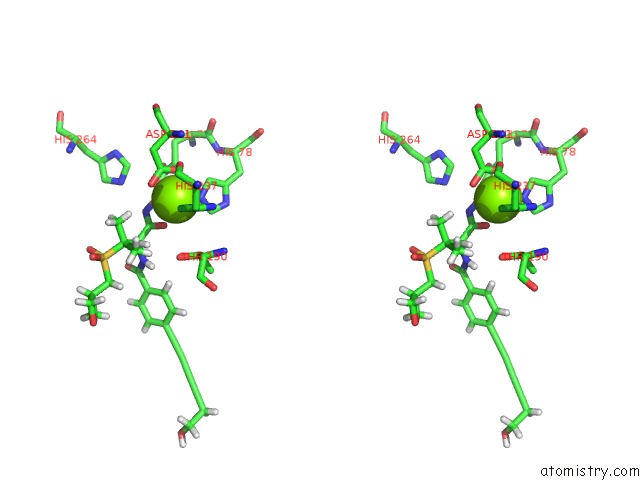
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 1 of Co-Crystal Structure of P. Aeruginosa Lpxc-50228 Complex within 5.0Å range:
|
Reference:
F.Cohen,
J.B.Aggen,
L.D.Andrews,
Z.Assar,
J.Boggs,
T.Choi,
P.Dozzo,
A.N.Easterday,
C.M.Haglund,
D.J.Hildebrandt,
M.C.Holt,
K.Joly,
A.Jubb,
Z.Kamal,
T.R.Kane,
A.W.Konradi,
K.M.Krause,
M.S.Linsell,
T.D.Machajewski,
O.Miroshnikova,
H.E.Moser,
V.Nieto,
T.Phan,
C.Plato,
A.W.Serio,
J.Seroogy,
A.Shakhmin,
A.J.Stein,
A.D.Sun,
S.Sviridov,
Z.Wang,
K.Wlasichuk,
W.Yang,
X.Zhou,
H.Zhu,
R.T.Cirz.
Optimization of Lpxc Inhibitors For Antibacterial Activity and Cardiovascular Safety. Chemmedchem V. 14 1560 2019.
ISSN: ESSN 1860-7187
PubMed: 31283109
DOI: 10.1002/CMDC.201900287
Page generated: Tue Oct 1 11:55:57 2024
ISSN: ESSN 1860-7187
PubMed: 31283109
DOI: 10.1002/CMDC.201900287
Last articles
I in 6DOCI in 6DOB
I in 6DOA
I in 6DE8
I in 6DO9
I in 6DO8
I in 6DMV
I in 6D13
I in 6DFN
I in 6DMN