Magnesium »
PDB 6n7n-6nk8 »
6njh »
Magnesium in PDB 6njh: Crystal Structure of the PDE4D Catalytic Domain and UCR2 Regulatory Helix with T-48
Enzymatic activity of Crystal Structure of the PDE4D Catalytic Domain and UCR2 Regulatory Helix with T-48
All present enzymatic activity of Crystal Structure of the PDE4D Catalytic Domain and UCR2 Regulatory Helix with T-48:
3.1.4.53;
3.1.4.53;
Protein crystallography data
The structure of Crystal Structure of the PDE4D Catalytic Domain and UCR2 Regulatory Helix with T-48, PDB code: 6njh
was solved by
D.Fox Iii,
J.W.Fairman,
M.E.Gurney,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 19.91 / 2.15 |
| Space group | P 1 21 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 81.640, 82.070, 116.530, 90.00, 110.23, 90.00 |
| R / Rfree (%) | 17.2 / 21 |
Other elements in 6njh:
The structure of Crystal Structure of the PDE4D Catalytic Domain and UCR2 Regulatory Helix with T-48 also contains other interesting chemical elements:
| Chlorine | (Cl) | 4 atoms |
| Zinc | (Zn) | 4 atoms |
Magnesium Binding Sites:
The binding sites of Magnesium atom in the Crystal Structure of the PDE4D Catalytic Domain and UCR2 Regulatory Helix with T-48
(pdb code 6njh). This binding sites where shown within
5.0 Angstroms radius around Magnesium atom.
In total 6 binding sites of Magnesium where determined in the Crystal Structure of the PDE4D Catalytic Domain and UCR2 Regulatory Helix with T-48, PDB code: 6njh:
Jump to Magnesium binding site number: 1; 2; 3; 4; 5; 6;
In total 6 binding sites of Magnesium where determined in the Crystal Structure of the PDE4D Catalytic Domain and UCR2 Regulatory Helix with T-48, PDB code: 6njh:
Jump to Magnesium binding site number: 1; 2; 3; 4; 5; 6;
Magnesium binding site 1 out of 6 in 6njh
Go back to
Magnesium binding site 1 out
of 6 in the Crystal Structure of the PDE4D Catalytic Domain and UCR2 Regulatory Helix with T-48
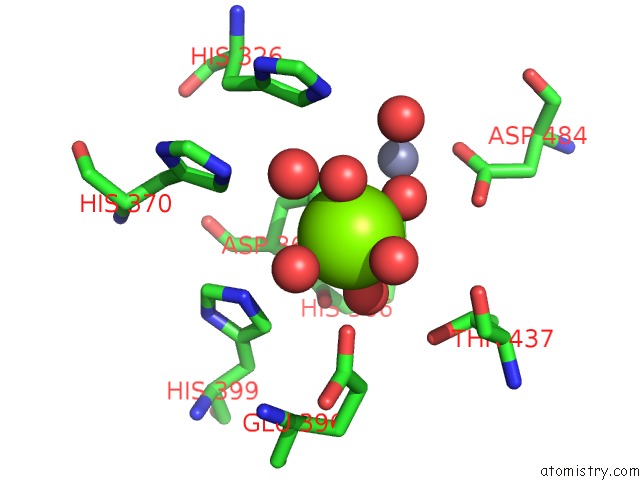
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 1 of Crystal Structure of the PDE4D Catalytic Domain and UCR2 Regulatory Helix with T-48 within 5.0Å range:
|
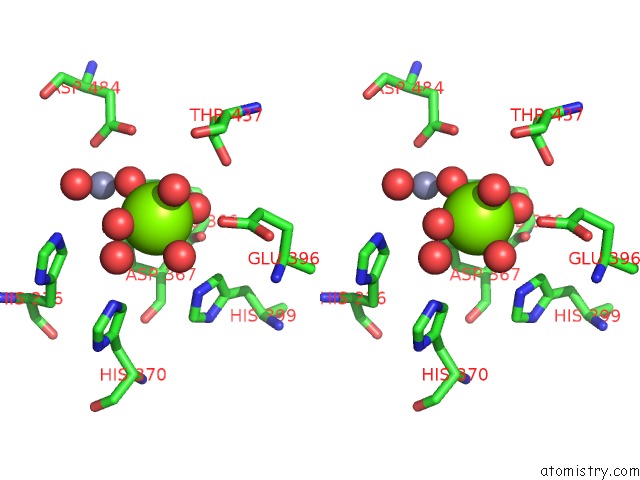
Magnesium binding site 2 out of 6 in 6njh
Go back to
Magnesium binding site 2 out
of 6 in the Crystal Structure of the PDE4D Catalytic Domain and UCR2 Regulatory Helix with T-48

Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 2 of Crystal Structure of the PDE4D Catalytic Domain and UCR2 Regulatory Helix with T-48 within 5.0Å range:
|
Magnesium binding site 3 out of 6 in 6njh
Go back to
Magnesium binding site 3 out
of 6 in the Crystal Structure of the PDE4D Catalytic Domain and UCR2 Regulatory Helix with T-48

Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 3 of Crystal Structure of the PDE4D Catalytic Domain and UCR2 Regulatory Helix with T-48 within 5.0Å range:
|
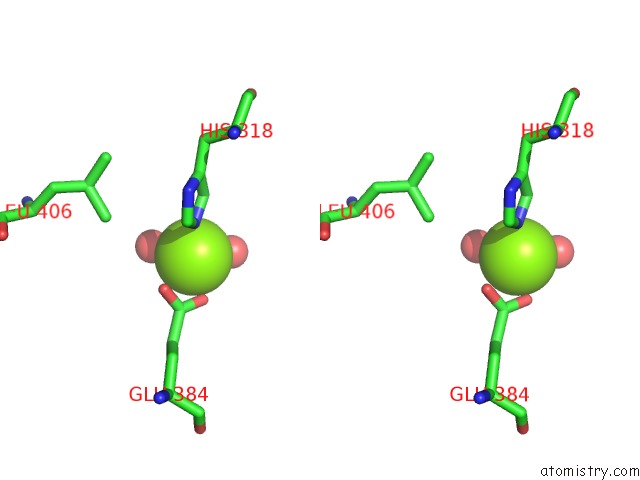
Magnesium binding site 4 out of 6 in 6njh
Go back to
Magnesium binding site 4 out
of 6 in the Crystal Structure of the PDE4D Catalytic Domain and UCR2 Regulatory Helix with T-48

Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 4 of Crystal Structure of the PDE4D Catalytic Domain and UCR2 Regulatory Helix with T-48 within 5.0Å range:
|
Magnesium binding site 5 out of 6 in 6njh
Go back to
Magnesium binding site 5 out
of 6 in the Crystal Structure of the PDE4D Catalytic Domain and UCR2 Regulatory Helix with T-48

Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 5 of Crystal Structure of the PDE4D Catalytic Domain and UCR2 Regulatory Helix with T-48 within 5.0Å range:
|
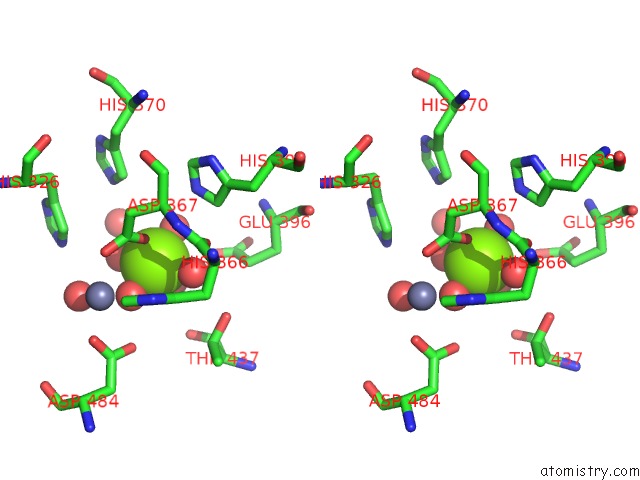
Magnesium binding site 6 out of 6 in 6njh
Go back to
Magnesium binding site 6 out
of 6 in the Crystal Structure of the PDE4D Catalytic Domain and UCR2 Regulatory Helix with T-48

Mono view
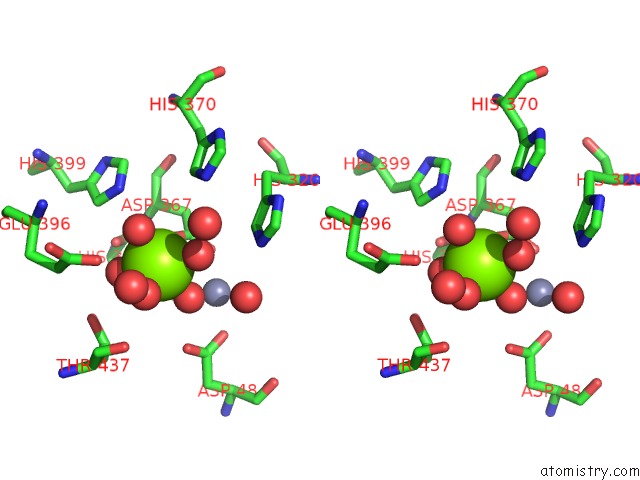
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 6 of Crystal Structure of the PDE4D Catalytic Domain and UCR2 Regulatory Helix with T-48 within 5.0Å range:
|
Reference:
M.E.Gurney,
R.A.Nugent,
X.Mo,
J.A.Sindac,
T.J.Hagen,
D.Fox Iii,
J.M.O'donnell,
C.Zhang,
Y.Xu,
H.T.Zhang,
V.E.Groppi,
M.Bailie,
R.E.White,
D.L.Romero,
A.S.Vellekoop,
J.R.Walker,
M.D.Surman,
L.Zhu,
R.F.Campbell.
Design and Synthesis of Selective Phosphodiesterase 4D (PDE4D) Allosteric Inhibitors For the Treatment of Fragile X Syndrome and Other Brain Disorders. J.Med.Chem. V. 62 4884 2019.
ISSN: ISSN 0022-2623
PubMed: 31013090
DOI: 10.1021/ACS.JMEDCHEM.9B00193
Page generated: Tue Oct 1 12:40:52 2024
ISSN: ISSN 0022-2623
PubMed: 31013090
DOI: 10.1021/ACS.JMEDCHEM.9B00193
Last articles
I in 4S22I in 4S2H
I in 4QX5
I in 4S2G
I in 4S2F
I in 4RX1
I in 4S2D
I in 4RYM
I in 4REC
I in 4RCF