Magnesium »
PDB 6yxu-6z9h »
6z5u »
Magnesium in PDB 6z5u: Cryo-Em Structure of the A. Baumannii Mlabdef Complex Bound to Appnhp
Magnesium Binding Sites:
The binding sites of Magnesium atom in the Cryo-Em Structure of the A. Baumannii Mlabdef Complex Bound to Appnhp
(pdb code 6z5u). This binding sites where shown within
5.0 Angstroms radius around Magnesium atom.
In total 2 binding sites of Magnesium where determined in the Cryo-Em Structure of the A. Baumannii Mlabdef Complex Bound to Appnhp, PDB code: 6z5u:
Jump to Magnesium binding site number: 1; 2;
In total 2 binding sites of Magnesium where determined in the Cryo-Em Structure of the A. Baumannii Mlabdef Complex Bound to Appnhp, PDB code: 6z5u:
Jump to Magnesium binding site number: 1; 2;
Magnesium binding site 1 out of 2 in 6z5u
Go back to
Magnesium binding site 1 out
of 2 in the Cryo-Em Structure of the A. Baumannii Mlabdef Complex Bound to Appnhp

Mono view
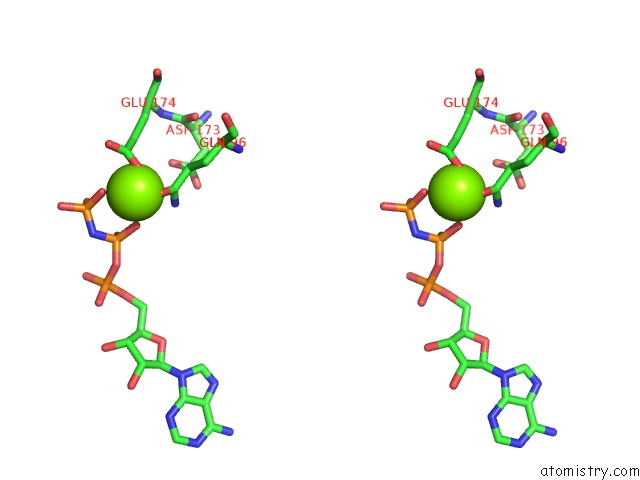
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 1 of Cryo-Em Structure of the A. Baumannii Mlabdef Complex Bound to Appnhp within 5.0Å range:
|
Magnesium binding site 2 out of 2 in 6z5u
Go back to
Magnesium binding site 2 out
of 2 in the Cryo-Em Structure of the A. Baumannii Mlabdef Complex Bound to Appnhp

Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 2 of Cryo-Em Structure of the A. Baumannii Mlabdef Complex Bound to Appnhp within 5.0Å range:
|
Reference:
D.Mann,
J.Fan,
D.P.Farrell,
K.Somboon,
A.Muenks,
S.B.Tzokov,
S.Khalid,
F.Dimaio,
S.I.Miller,
J.R.C.Bergeron.
Structure and Lipid Dynamics in the A. Baumannii Maintenance of Lipid Asymmetry (Mla) Inner Membrane Complex Biorxiv 2020.
DOI: 10.1101/2020.05.30.125013
Page generated: Wed Oct 2 02:05:34 2024
DOI: 10.1101/2020.05.30.125013
Last articles
Mg in 6KCCMg in 6KCA
Mg in 6KC0
Mg in 6KBZ
Mg in 6KAL
Mg in 6KAK
Mg in 6KBD
Mg in 6K9V
Mg in 6K83
Mg in 6K8K