Magnesium »
PDB 6zio-6zu9 »
6zqt »
Magnesium in PDB 6zqt: Crystal Structure of the RLIP76 Ral Binding Domain Mutant (E427H/Q433L/K440R) in Complex with Ralb-Gmppnp
Protein crystallography data
The structure of Crystal Structure of the RLIP76 Ral Binding Domain Mutant (E427H/Q433L/K440R) in Complex with Ralb-Gmppnp, PDB code: 6zqt
was solved by
C.Hurd,
P.Brear,
J.Revell,
S.Ross,
H.Mott,
D.Owen,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 50.40 / 1.51 |
| Space group | P 1 21 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 47.450, 77.430, 66.400, 90.00, 90.31, 90.00 |
| R / Rfree (%) | 18.9 / 22.1 |
Magnesium Binding Sites:
The binding sites of Magnesium atom in the Crystal Structure of the RLIP76 Ral Binding Domain Mutant (E427H/Q433L/K440R) in Complex with Ralb-Gmppnp
(pdb code 6zqt). This binding sites where shown within
5.0 Angstroms radius around Magnesium atom.
In total 2 binding sites of Magnesium where determined in the Crystal Structure of the RLIP76 Ral Binding Domain Mutant (E427H/Q433L/K440R) in Complex with Ralb-Gmppnp, PDB code: 6zqt:
Jump to Magnesium binding site number: 1; 2;
In total 2 binding sites of Magnesium where determined in the Crystal Structure of the RLIP76 Ral Binding Domain Mutant (E427H/Q433L/K440R) in Complex with Ralb-Gmppnp, PDB code: 6zqt:
Jump to Magnesium binding site number: 1; 2;
Magnesium binding site 1 out of 2 in 6zqt
Go back to
Magnesium binding site 1 out
of 2 in the Crystal Structure of the RLIP76 Ral Binding Domain Mutant (E427H/Q433L/K440R) in Complex with Ralb-Gmppnp
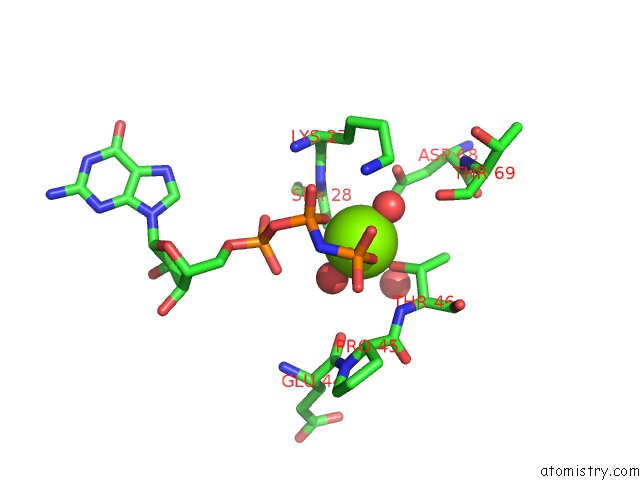
Mono view
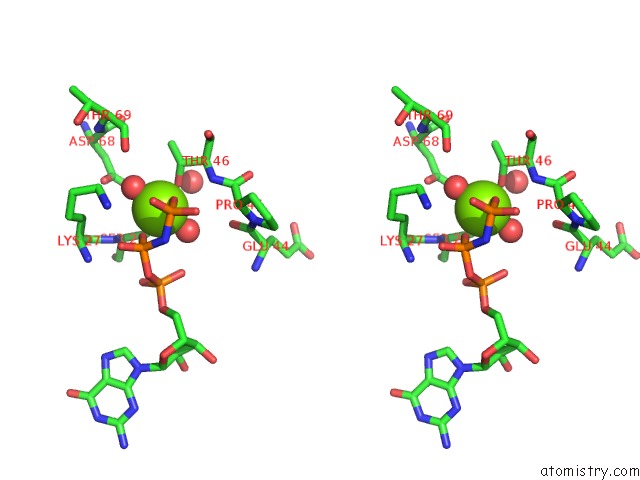
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 1 of Crystal Structure of the RLIP76 Ral Binding Domain Mutant (E427H/Q433L/K440R) in Complex with Ralb-Gmppnp within 5.0Å range:
|
Magnesium binding site 2 out of 2 in 6zqt
Go back to
Magnesium binding site 2 out
of 2 in the Crystal Structure of the RLIP76 Ral Binding Domain Mutant (E427H/Q433L/K440R) in Complex with Ralb-Gmppnp
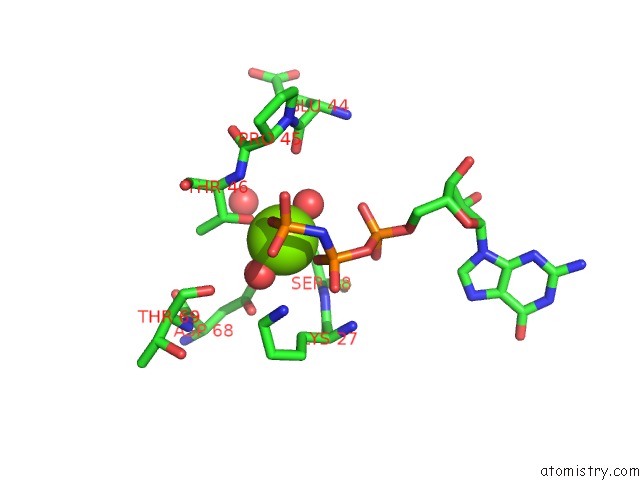
Mono view
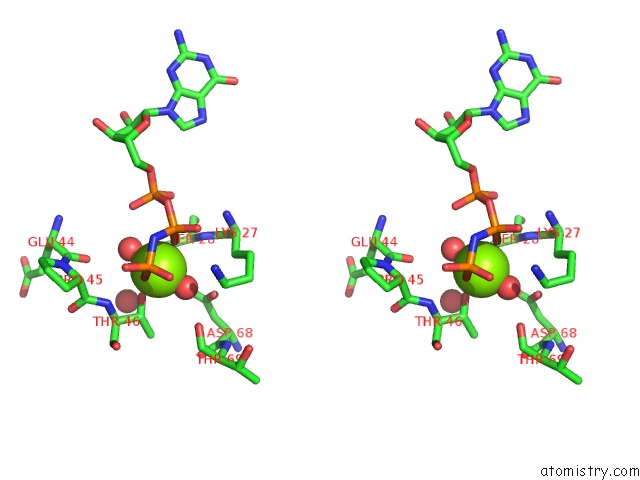
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 2 of Crystal Structure of the RLIP76 Ral Binding Domain Mutant (E427H/Q433L/K440R) in Complex with Ralb-Gmppnp within 5.0Å range:
|
Reference:
C.A.Hurd,
P.Brear,
J.Revell,
S.Ross,
H.R.Mott,
D.Owen.
Affinity Maturation of the RLIP76 Ral Binding Domain to Inform the Design of Stapled Peptides Targeting the Ral Gtpases. J.Biol.Chem. 2020.
ISSN: ESSN 1083-351X
PubMed: 33214225
DOI: 10.1074/JBC.RA120.015735
Page generated: Wed Oct 2 02:35:09 2024
ISSN: ESSN 1083-351X
PubMed: 33214225
DOI: 10.1074/JBC.RA120.015735
Last articles
I in 5KIOI in 5KIM
I in 5K6U
I in 5KC1
I in 5K7P
I in 5JVF
I in 5JSI
I in 5K27
I in 5JRF
I in 5IQY