Magnesium »
PDB 7mir-7mnz »
7mkd »
Magnesium in PDB 7mkd: Cryo-Em Structure of Escherichia Coli Rna Polymerase Bound to Lambda Pr Promoter Dna (Class 1)
Enzymatic activity of Cryo-Em Structure of Escherichia Coli Rna Polymerase Bound to Lambda Pr Promoter Dna (Class 1)
All present enzymatic activity of Cryo-Em Structure of Escherichia Coli Rna Polymerase Bound to Lambda Pr Promoter Dna (Class 1):
2.7.7.6;
2.7.7.6;
Other elements in 7mkd:
The structure of Cryo-Em Structure of Escherichia Coli Rna Polymerase Bound to Lambda Pr Promoter Dna (Class 1) also contains other interesting chemical elements:
| Zinc | (Zn) | 2 atoms |
Magnesium Binding Sites:
The binding sites of Magnesium atom in the Cryo-Em Structure of Escherichia Coli Rna Polymerase Bound to Lambda Pr Promoter Dna (Class 1)
(pdb code 7mkd). This binding sites where shown within
5.0 Angstroms radius around Magnesium atom.
In total only one binding site of Magnesium was determined in the Cryo-Em Structure of Escherichia Coli Rna Polymerase Bound to Lambda Pr Promoter Dna (Class 1), PDB code: 7mkd:
In total only one binding site of Magnesium was determined in the Cryo-Em Structure of Escherichia Coli Rna Polymerase Bound to Lambda Pr Promoter Dna (Class 1), PDB code: 7mkd:
Magnesium binding site 1 out of 1 in 7mkd
Go back to
Magnesium binding site 1 out
of 1 in the Cryo-Em Structure of Escherichia Coli Rna Polymerase Bound to Lambda Pr Promoter Dna (Class 1)
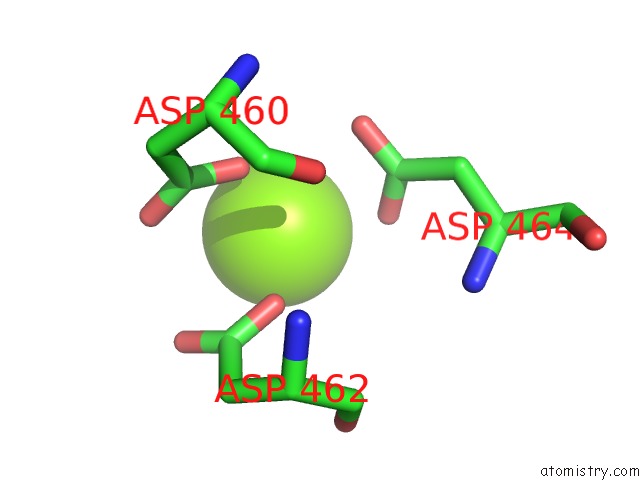
Mono view
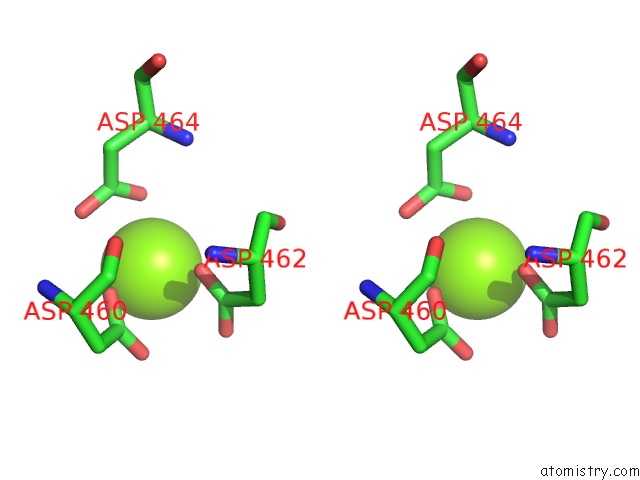
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 1 of Cryo-Em Structure of Escherichia Coli Rna Polymerase Bound to Lambda Pr Promoter Dna (Class 1) within 5.0Å range:
|
Reference:
R.M.Saecker,
J.Chen,
C.Chiu,
B.Malone,
J.Sotiris,
M.Ebrahim,
L.Y.Yen,
E.T.Eng,
S.A.Darst.
Structural Origins of Escherichia Coli Rna Polymerase Open Promoter Complex Stability To Be Published.
Page generated: Thu Oct 3 01:00:31 2024
Last articles
Fe in 2YXOFe in 2YRS
Fe in 2YXC
Fe in 2YNM
Fe in 2YVJ
Fe in 2YP1
Fe in 2YU2
Fe in 2YU1
Fe in 2YQB
Fe in 2YOO