Magnesium »
PDB 7o11-7o6a »
7o4j »
Magnesium in PDB 7o4j: Yeast Rna Polymerase II Transcription Pre-Initiation Complex (Consensus)
Enzymatic activity of Yeast Rna Polymerase II Transcription Pre-Initiation Complex (Consensus)
All present enzymatic activity of Yeast Rna Polymerase II Transcription Pre-Initiation Complex (Consensus):
2.7.7.6; 3.6.4.12;
2.7.7.6; 3.6.4.12;
Other elements in 7o4j:
The structure of Yeast Rna Polymerase II Transcription Pre-Initiation Complex (Consensus) also contains other interesting chemical elements:
| Zinc | (Zn) | 17 atoms |
| Iron | (Fe) | 4 atoms |
| Fluorine | (F) | 3 atoms |
Magnesium Binding Sites:
The binding sites of Magnesium atom in the Yeast Rna Polymerase II Transcription Pre-Initiation Complex (Consensus)
(pdb code 7o4j). This binding sites where shown within
5.0 Angstroms radius around Magnesium atom.
In total 2 binding sites of Magnesium where determined in the Yeast Rna Polymerase II Transcription Pre-Initiation Complex (Consensus), PDB code: 7o4j:
Jump to Magnesium binding site number: 1; 2;
In total 2 binding sites of Magnesium where determined in the Yeast Rna Polymerase II Transcription Pre-Initiation Complex (Consensus), PDB code: 7o4j:
Jump to Magnesium binding site number: 1; 2;
Magnesium binding site 1 out of 2 in 7o4j
Go back to
Magnesium binding site 1 out
of 2 in the Yeast Rna Polymerase II Transcription Pre-Initiation Complex (Consensus)
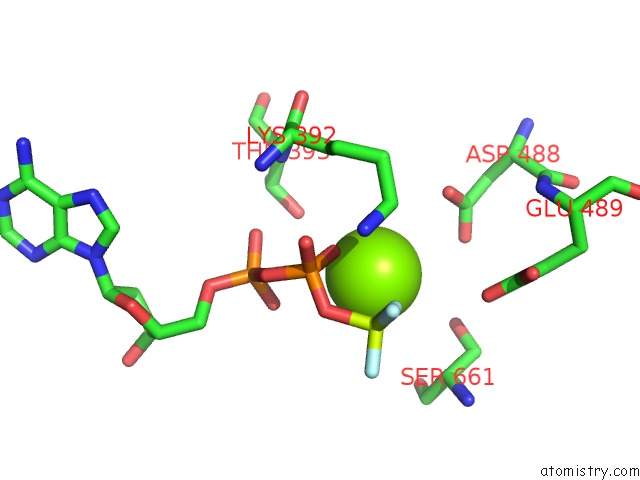
Mono view
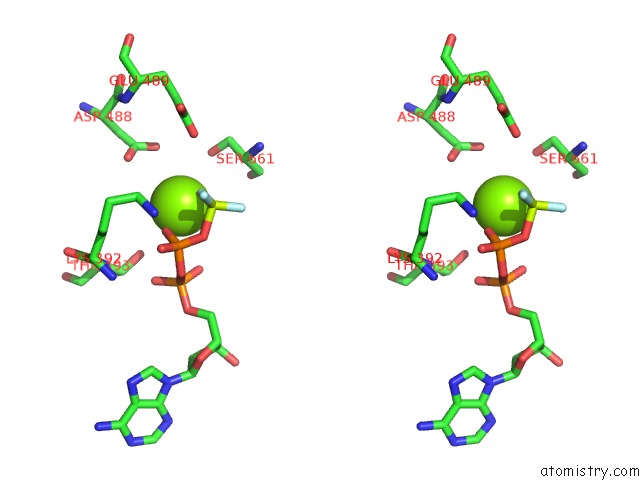
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 1 of Yeast Rna Polymerase II Transcription Pre-Initiation Complex (Consensus) within 5.0Å range:
|
Magnesium binding site 2 out of 2 in 7o4j
Go back to
Magnesium binding site 2 out
of 2 in the Yeast Rna Polymerase II Transcription Pre-Initiation Complex (Consensus)
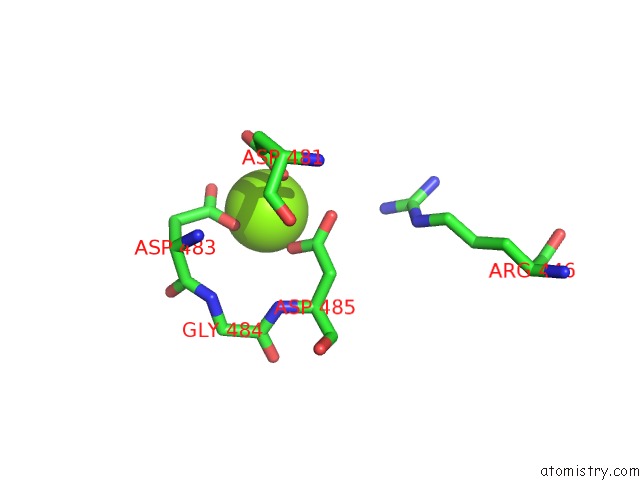
Mono view
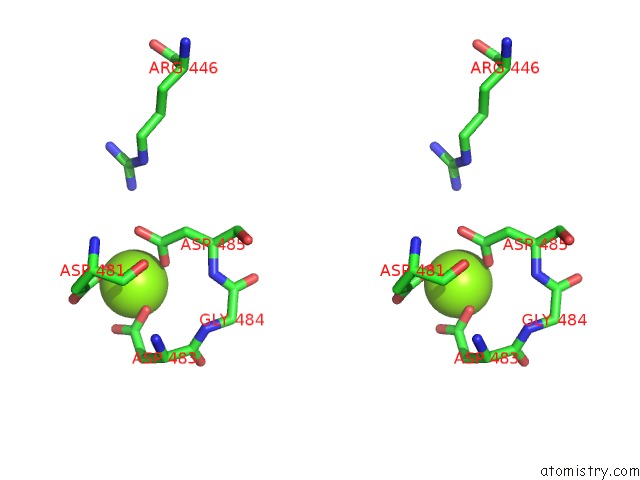
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 2 of Yeast Rna Polymerase II Transcription Pre-Initiation Complex (Consensus) within 5.0Å range:
|
Reference:
S.Schilbach,
S.Aibara,
C.Dienemann,
F.Grabbe,
P.Cramer.
Structure of Rna Polymerase II Pre-Initiation Complex at 2.9 Angstrom Defines Initial Dna Opening Cell(Cambridge,Mass.) 2021.
ISSN: ISSN 0092-8674
DOI: 10.1016/J.CELL.2021.05.012
Page generated: Thu Oct 3 02:32:44 2024
ISSN: ISSN 0092-8674
DOI: 10.1016/J.CELL.2021.05.012
Last articles
K in 2GJBK in 2GIX
K in 2GHK
K in 2GHS
K in 2GDI
K in 2GHH
K in 2G6P
K in 2G4V
K in 2G4N
K in 2FZW