Magnesium »
PDB 7o6c-7ocq »
7oar »
Magnesium in PDB 7oar: Crystal Structure of Helicase PIF1 From Thermus Oshimai in Complex with Parallel G-Quadruplex
Protein crystallography data
The structure of Crystal Structure of Helicase PIF1 From Thermus Oshimai in Complex with Parallel G-Quadruplex, PDB code: 7oar
was solved by
Y.X.Dai,
N.N.Liu,
H.L.Guo,
W.F.Chen,
S.Rety,
X.G.Xi,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 37.39 / 2.58 |
| Space group | P 62 2 2 |
| Cell size a, b, c (Å), α, β, γ (°) | 151.782, 151.782, 219.534, 90, 90, 120 |
| R / Rfree (%) | 19.4 / 24.3 |
Other elements in 7oar:
The structure of Crystal Structure of Helicase PIF1 From Thermus Oshimai in Complex with Parallel G-Quadruplex also contains other interesting chemical elements:
| Aluminium | (Al) | 2 atoms |
| Potassium | (K) | 3 atoms |
| Fluorine | (F) | 8 atoms |
Magnesium Binding Sites:
The binding sites of Magnesium atom in the Crystal Structure of Helicase PIF1 From Thermus Oshimai in Complex with Parallel G-Quadruplex
(pdb code 7oar). This binding sites where shown within
5.0 Angstroms radius around Magnesium atom.
In total 2 binding sites of Magnesium where determined in the Crystal Structure of Helicase PIF1 From Thermus Oshimai in Complex with Parallel G-Quadruplex, PDB code: 7oar:
Jump to Magnesium binding site number: 1; 2;
In total 2 binding sites of Magnesium where determined in the Crystal Structure of Helicase PIF1 From Thermus Oshimai in Complex with Parallel G-Quadruplex, PDB code: 7oar:
Jump to Magnesium binding site number: 1; 2;
Magnesium binding site 1 out of 2 in 7oar
Go back to
Magnesium binding site 1 out
of 2 in the Crystal Structure of Helicase PIF1 From Thermus Oshimai in Complex with Parallel G-Quadruplex
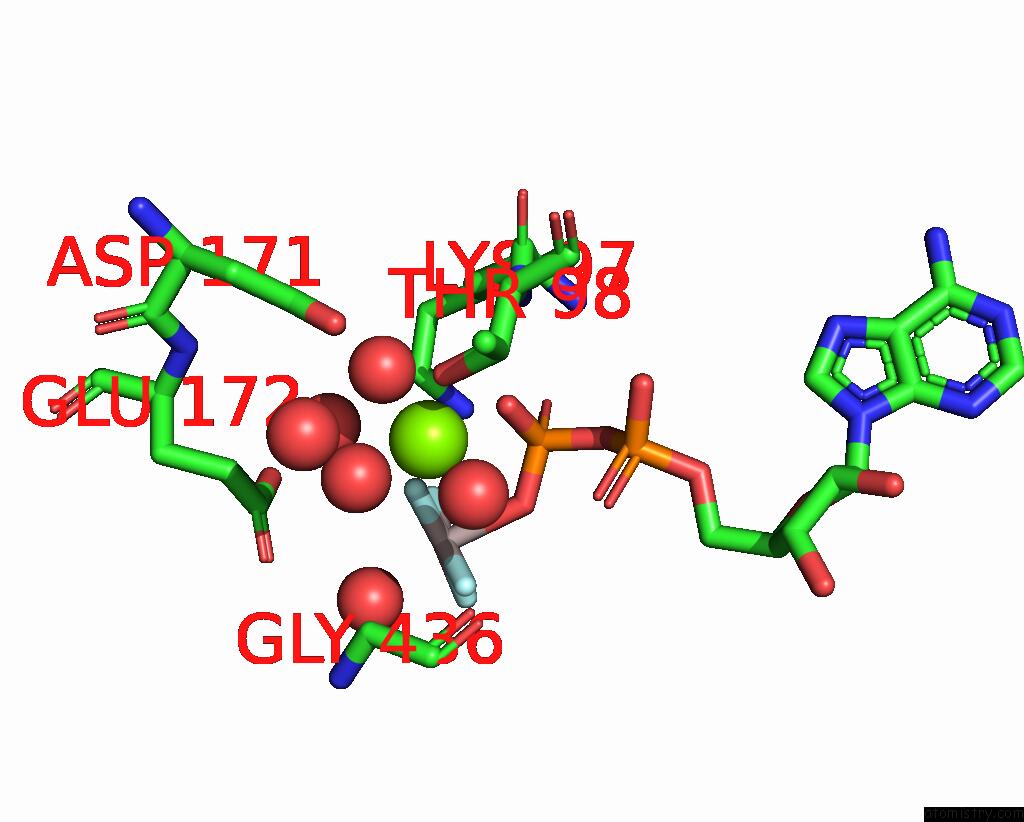
Mono view
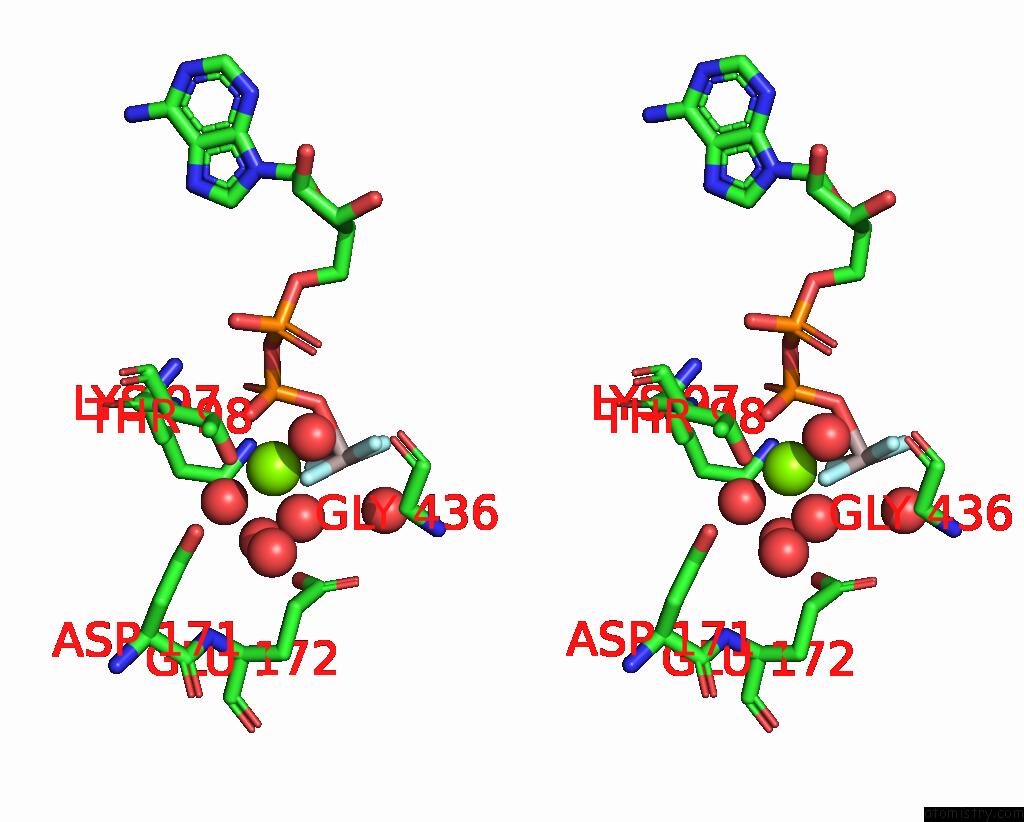
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 1 of Crystal Structure of Helicase PIF1 From Thermus Oshimai in Complex with Parallel G-Quadruplex within 5.0Å range:
|
Magnesium binding site 2 out of 2 in 7oar
Go back to
Magnesium binding site 2 out
of 2 in the Crystal Structure of Helicase PIF1 From Thermus Oshimai in Complex with Parallel G-Quadruplex
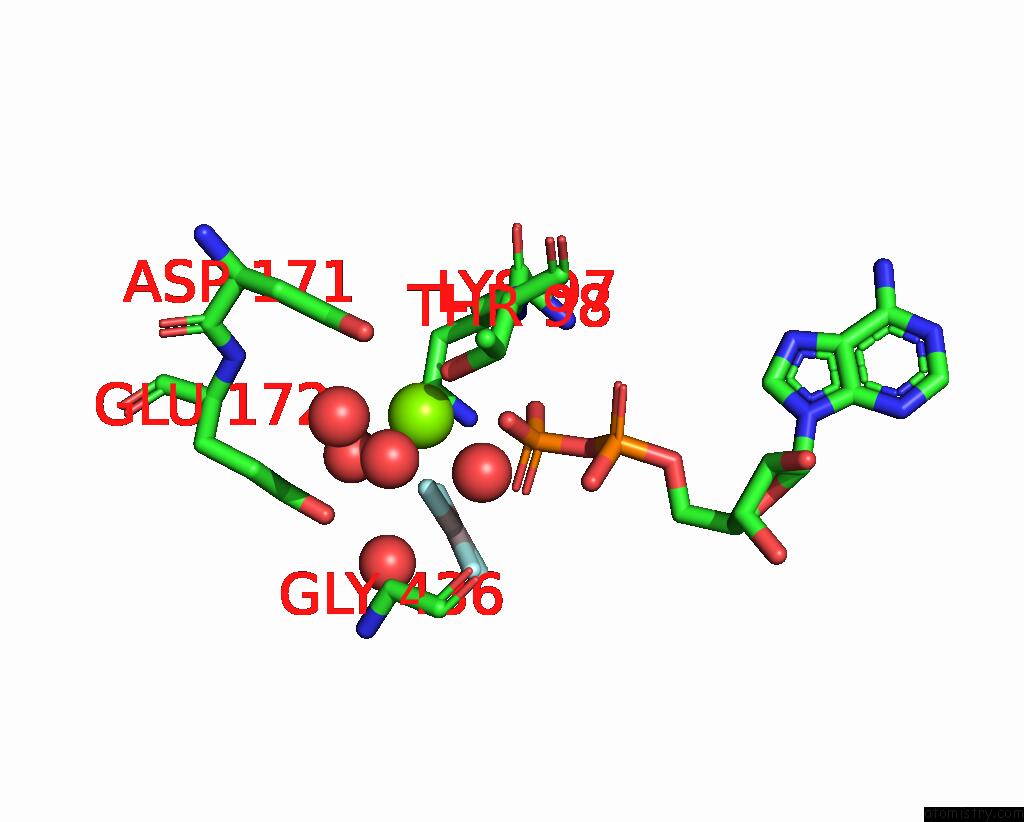
Mono view
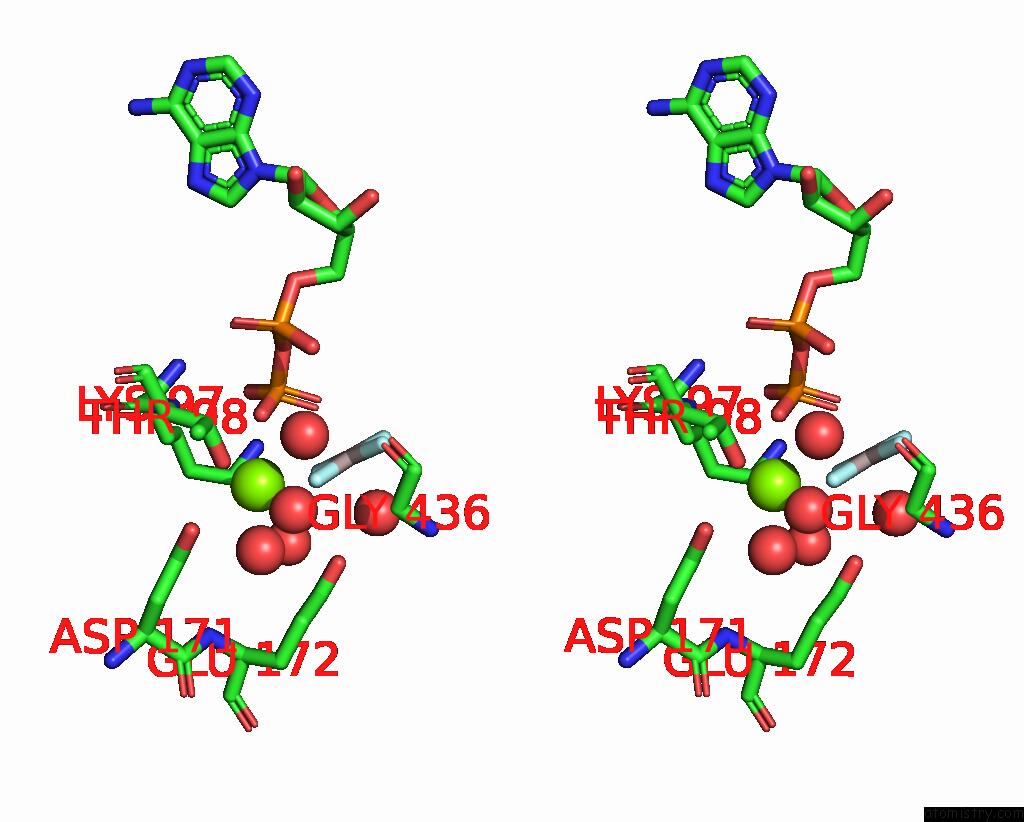
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 2 of Crystal Structure of Helicase PIF1 From Thermus Oshimai in Complex with Parallel G-Quadruplex within 5.0Å range:
|
Reference:
Y.X.Dai,
H.L.Guo,
N.N.Liu,
W.F.Chen,
X.Ai,
H.H.Li,
B.Sun,
X.M.Hou,
S.Rety,
X.G.Xi.
Structural Mechanism Underpinning Thermus Oshimai PIF1-Mediated G-Quadruplex Unfolding. Embo Rep. V. 23 53874 2022.
ISSN: ESSN 1469-3178
PubMed: 35736675
DOI: 10.15252/EMBR.202153874
Page generated: Thu Oct 3 02:52:00 2024
ISSN: ESSN 1469-3178
PubMed: 35736675
DOI: 10.15252/EMBR.202153874
Last articles
I in 3RX6I in 3RSI
I in 3RG2
I in 3RTN
I in 3RTM
I in 3RTH
I in 3RSX
I in 3RKE
I in 3RSV
I in 3PFD