Magnesium »
PDB 7ocr-7oim »
7oh4 »
Magnesium in PDB 7oh4: Cryo-Em Structure of DRS2P-CDC50P in the E1 State with PI4P and MG2+ Bound
Enzymatic activity of Cryo-Em Structure of DRS2P-CDC50P in the E1 State with PI4P and MG2+ Bound
All present enzymatic activity of Cryo-Em Structure of DRS2P-CDC50P in the E1 State with PI4P and MG2+ Bound:
7.6.2.1;
7.6.2.1;
Magnesium Binding Sites:
The binding sites of Magnesium atom in the Cryo-Em Structure of DRS2P-CDC50P in the E1 State with PI4P and MG2+ Bound
(pdb code 7oh4). This binding sites where shown within
5.0 Angstroms radius around Magnesium atom.
In total only one binding site of Magnesium was determined in the Cryo-Em Structure of DRS2P-CDC50P in the E1 State with PI4P and MG2+ Bound, PDB code: 7oh4:
In total only one binding site of Magnesium was determined in the Cryo-Em Structure of DRS2P-CDC50P in the E1 State with PI4P and MG2+ Bound, PDB code: 7oh4:
Magnesium binding site 1 out of 1 in 7oh4
Go back to
Magnesium binding site 1 out
of 1 in the Cryo-Em Structure of DRS2P-CDC50P in the E1 State with PI4P and MG2+ Bound
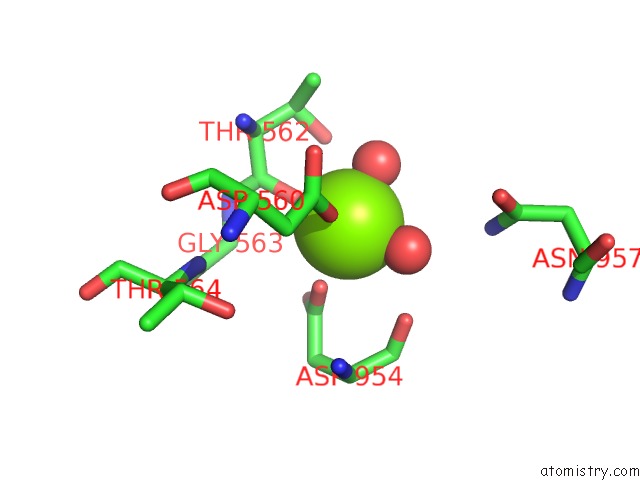
Mono view
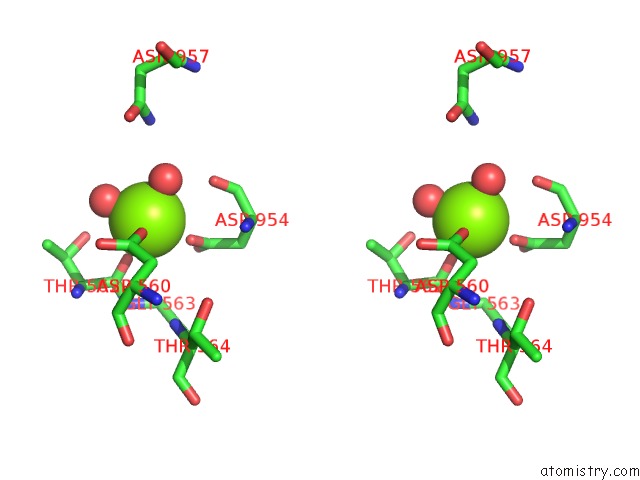
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 1 of Cryo-Em Structure of DRS2P-CDC50P in the E1 State with PI4P and MG2+ Bound within 5.0Å range:
|
Reference:
M.Timcenko,
T.Dieudonne,
C.Montigny,
T.Boesen,
J.A.Lyons,
G.Lenoir,
P.Nissen.
Structural Basis of Substrate-Independent Phosphorylation in A P4-Atpase Lipid Flippase J.Mol.Biol. 67062 2021.
ISSN: ESSN 1089-8638
PubMed: 34023399
DOI: 10.1016/J.JMB.2021.167062
Page generated: Thu Oct 3 02:58:11 2024
ISSN: ESSN 1089-8638
PubMed: 34023399
DOI: 10.1016/J.JMB.2021.167062
Last articles
I in 5K2XI in 5KIO
I in 5KIM
I in 5K6U
I in 5KC1
I in 5K7P
I in 5JVF
I in 5JSI
I in 5K27
I in 5JRF