Magnesium »
PDB 7py1-7q7j »
7q1u »
Magnesium in PDB 7q1u: Structure of Hedgehog Acyltransferase (Hhat) in Complex with Megabody 177 Bound to Non-Hydrolysable Palmitoyl-Coa (Composite Map)
Other elements in 7q1u:
The structure of Structure of Hedgehog Acyltransferase (Hhat) in Complex with Megabody 177 Bound to Non-Hydrolysable Palmitoyl-Coa (Composite Map) also contains other interesting chemical elements:
| Iron | (Fe) | 1 atom |
Magnesium Binding Sites:
The binding sites of Magnesium atom in the Structure of Hedgehog Acyltransferase (Hhat) in Complex with Megabody 177 Bound to Non-Hydrolysable Palmitoyl-Coa (Composite Map)
(pdb code 7q1u). This binding sites where shown within
5.0 Angstroms radius around Magnesium atom.
In total only one binding site of Magnesium was determined in the Structure of Hedgehog Acyltransferase (Hhat) in Complex with Megabody 177 Bound to Non-Hydrolysable Palmitoyl-Coa (Composite Map), PDB code: 7q1u:
In total only one binding site of Magnesium was determined in the Structure of Hedgehog Acyltransferase (Hhat) in Complex with Megabody 177 Bound to Non-Hydrolysable Palmitoyl-Coa (Composite Map), PDB code: 7q1u:
Magnesium binding site 1 out of 1 in 7q1u
Go back to
Magnesium binding site 1 out
of 1 in the Structure of Hedgehog Acyltransferase (Hhat) in Complex with Megabody 177 Bound to Non-Hydrolysable Palmitoyl-Coa (Composite Map)
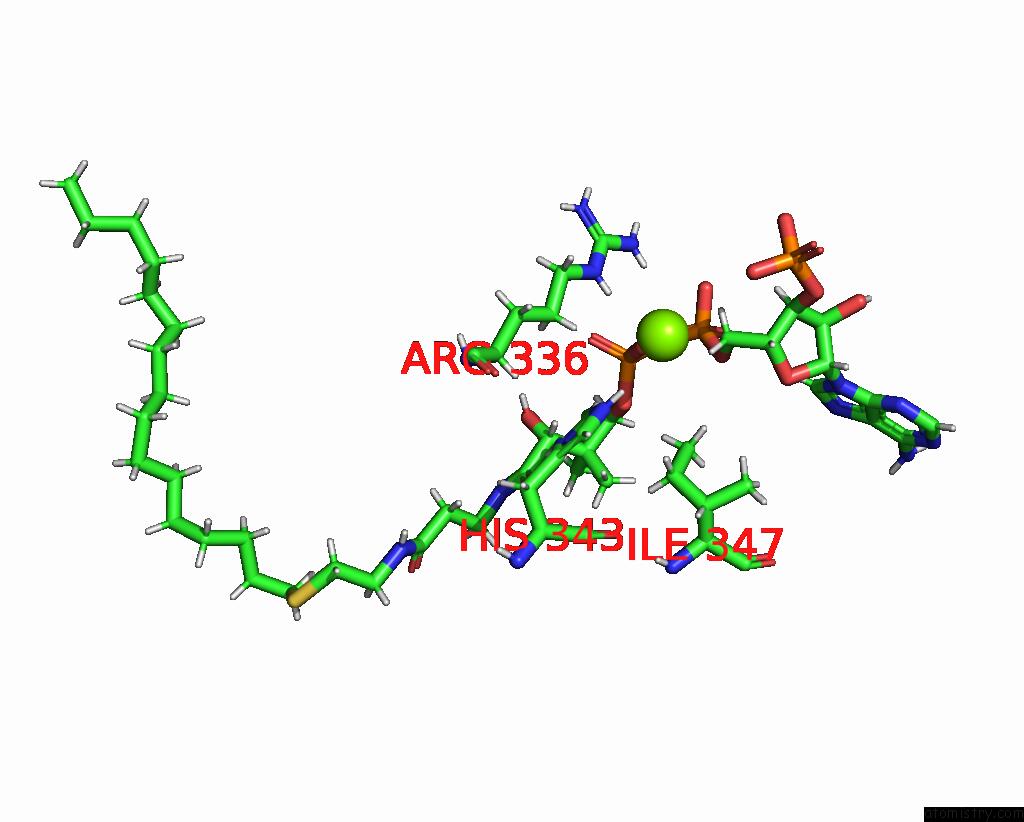
Mono view
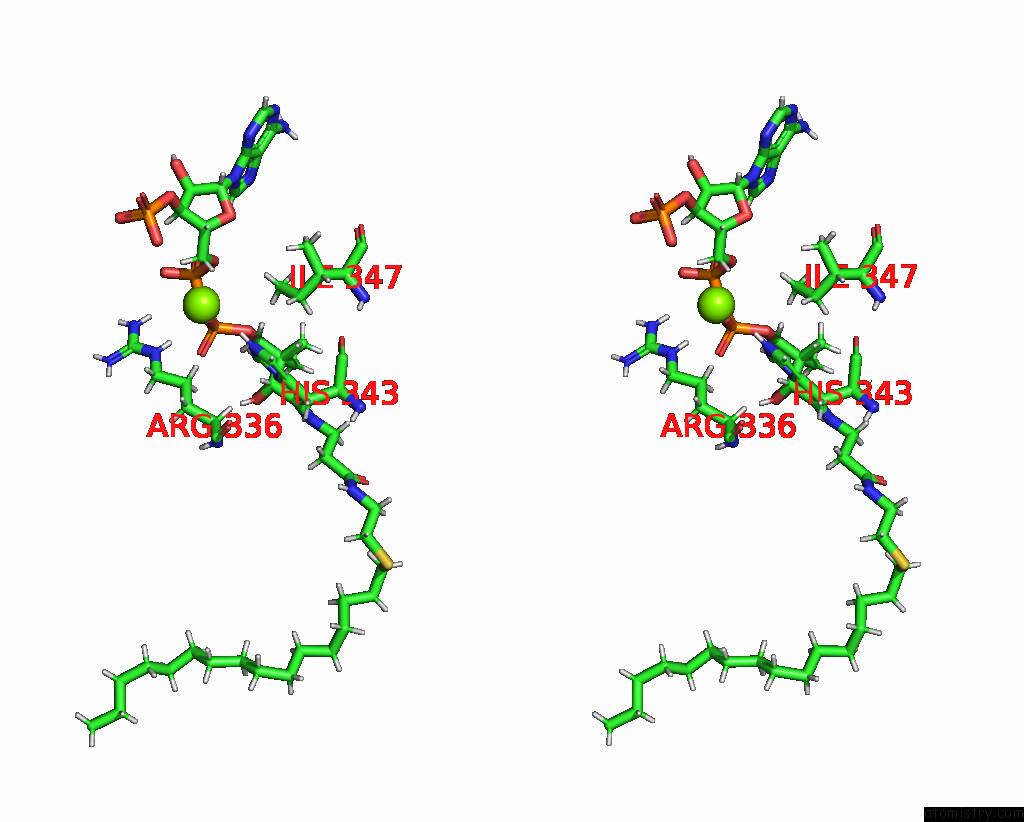
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 1 of Structure of Hedgehog Acyltransferase (Hhat) in Complex with Megabody 177 Bound to Non-Hydrolysable Palmitoyl-Coa (Composite Map) within 5.0Å range:
|
Reference:
C.E.Coupland,
S.A.Andrei,
T.B.Ansell,
L.Carrique,
P.Kumar,
L.Sefer,
R.A.Schwab,
E.F.X.Byrne,
E.Pardon,
J.Steyaert,
A.I.Magee,
T.Lanyon-Hogg,
M.S.P.Sansom,
E.W.Tate,
C.Siebold.
Structure, Mechanism, and Inhibition of Hedgehog Acyltransferase. Mol.Cell V. 81 5025 2021.
ISSN: ISSN 1097-2765
PubMed: 34890564
DOI: 10.1016/J.MOLCEL.2021.11.018
Page generated: Thu Oct 3 04:57:02 2024
ISSN: ISSN 1097-2765
PubMed: 34890564
DOI: 10.1016/J.MOLCEL.2021.11.018
Last articles
Mg in 3CB3Mg in 3CC6
Mg in 3C9U
Mg in 3CBT
Mg in 3CBQ
Mg in 3CBG
Mg in 3CBE
Mg in 3C9T
Mg in 3CB9
Mg in 3CAW