Magnesium »
PDB 8acg-8aom »
8age »
Magnesium in PDB 8age: Structure of Yeast Oligosaccharylransferase Complex with Acceptor Peptide Bound
Enzymatic activity of Structure of Yeast Oligosaccharylransferase Complex with Acceptor Peptide Bound
All present enzymatic activity of Structure of Yeast Oligosaccharylransferase Complex with Acceptor Peptide Bound:
2.4.99.18;
2.4.99.18;
Magnesium Binding Sites:
The binding sites of Magnesium atom in the Structure of Yeast Oligosaccharylransferase Complex with Acceptor Peptide Bound
(pdb code 8age). This binding sites where shown within
5.0 Angstroms radius around Magnesium atom.
In total only one binding site of Magnesium was determined in the Structure of Yeast Oligosaccharylransferase Complex with Acceptor Peptide Bound, PDB code: 8age:
In total only one binding site of Magnesium was determined in the Structure of Yeast Oligosaccharylransferase Complex with Acceptor Peptide Bound, PDB code: 8age:
Magnesium binding site 1 out of 1 in 8age
Go back to
Magnesium binding site 1 out
of 1 in the Structure of Yeast Oligosaccharylransferase Complex with Acceptor Peptide Bound
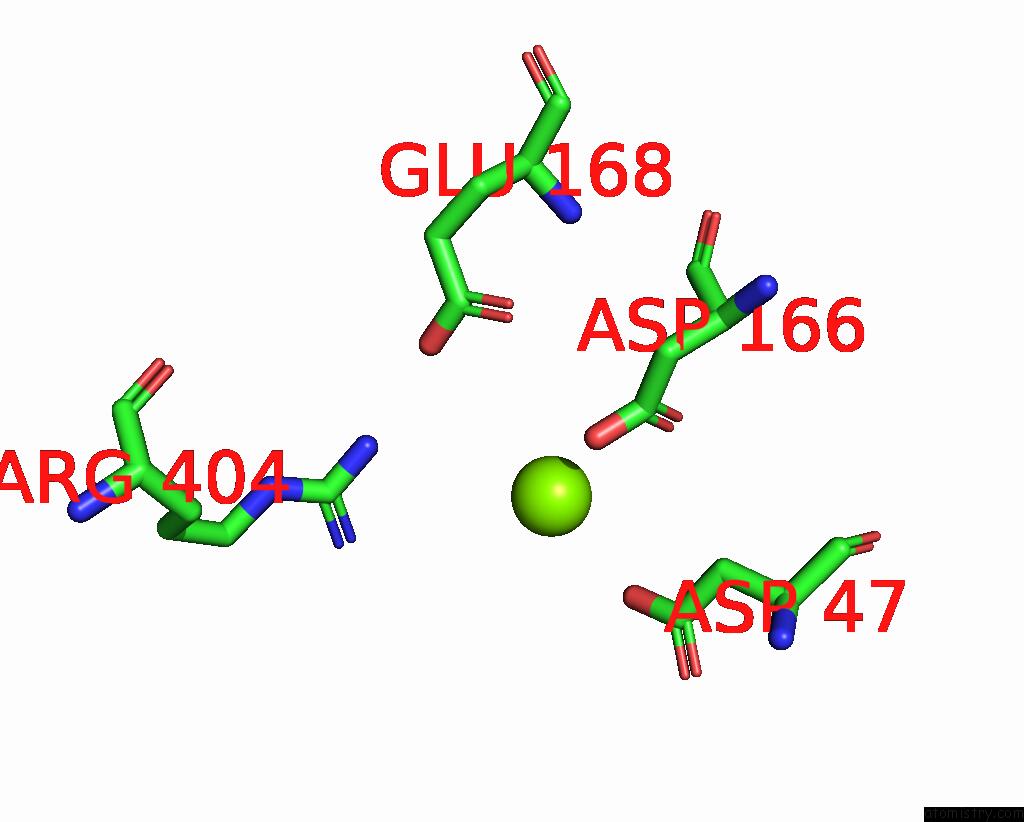
Mono view
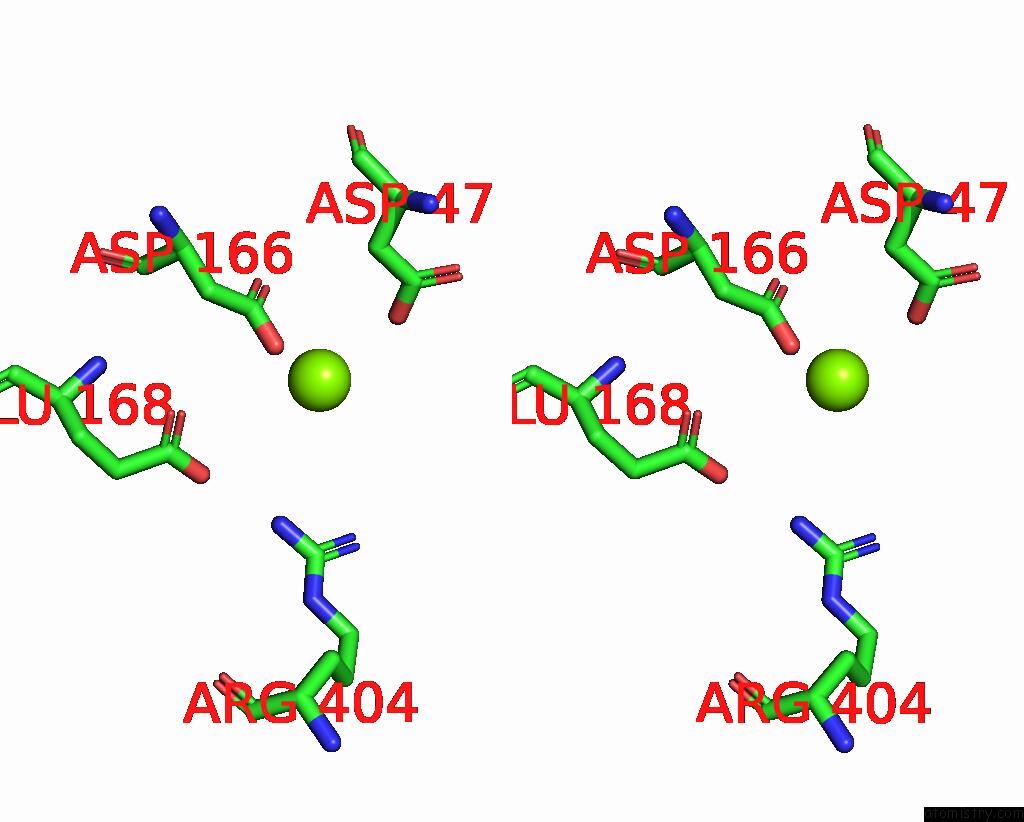
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 1 of Structure of Yeast Oligosaccharylransferase Complex with Acceptor Peptide Bound within 5.0Å range:
|
Reference:
A.S.Ramirez,
M.De Capitani,
G.Pesciullesi,
J.Kowal,
J.S.Bloch,
R.N.Irobalieva,
J.L.Reymond,
M.Aebi,
K.P.Locher.
Molecular Basis For Glycan Recognition and Reaction Priming of Eukaryotic Oligosaccharyltransferase. Nat Commun V. 13 7296 2022.
ISSN: ESSN 2041-1723
PubMed: 36435935
DOI: 10.1038/S41467-022-35067-X
Page generated: Thu Oct 3 18:07:03 2024
ISSN: ESSN 2041-1723
PubMed: 36435935
DOI: 10.1038/S41467-022-35067-X
Last articles
Mg in 7FURMg in 7FUJ
Mg in 7GQT
Mg in 7FUI
Mg in 7FTR
Mg in 7FTU
Mg in 7FUB
Mg in 7FSC
Mg in 7FSB
Mg in 7FSA