Magnesium »
PDB 8g4f-8ghu »
8g9u »
Magnesium in PDB 8g9u: Exploiting Activation and Inactivation Mechanisms in Type I-C Crispr- CAS3 For Genome Editing Applications
Magnesium Binding Sites:
The binding sites of Magnesium atom in the Exploiting Activation and Inactivation Mechanisms in Type I-C Crispr- CAS3 For Genome Editing Applications
(pdb code 8g9u). This binding sites where shown within
5.0 Angstroms radius around Magnesium atom.
In total 2 binding sites of Magnesium where determined in the Exploiting Activation and Inactivation Mechanisms in Type I-C Crispr- CAS3 For Genome Editing Applications, PDB code: 8g9u:
Jump to Magnesium binding site number: 1; 2;
In total 2 binding sites of Magnesium where determined in the Exploiting Activation and Inactivation Mechanisms in Type I-C Crispr- CAS3 For Genome Editing Applications, PDB code: 8g9u:
Jump to Magnesium binding site number: 1; 2;
Magnesium binding site 1 out of 2 in 8g9u
Go back to
Magnesium binding site 1 out
of 2 in the Exploiting Activation and Inactivation Mechanisms in Type I-C Crispr- CAS3 For Genome Editing Applications
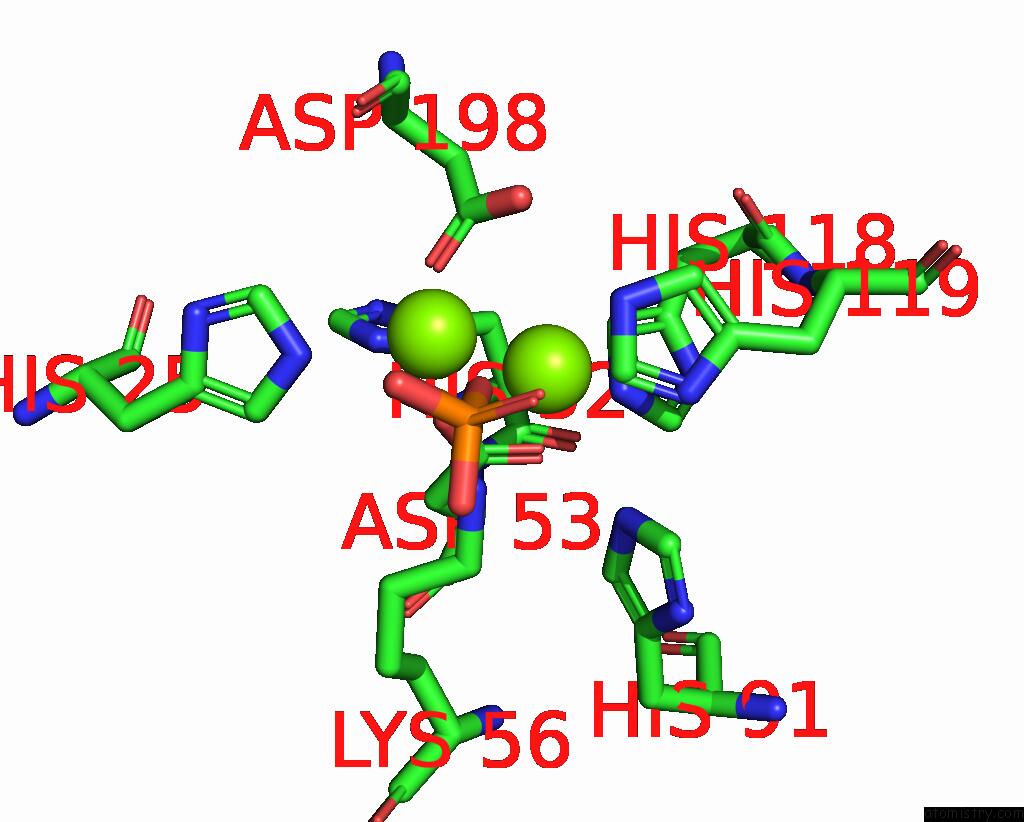
Mono view
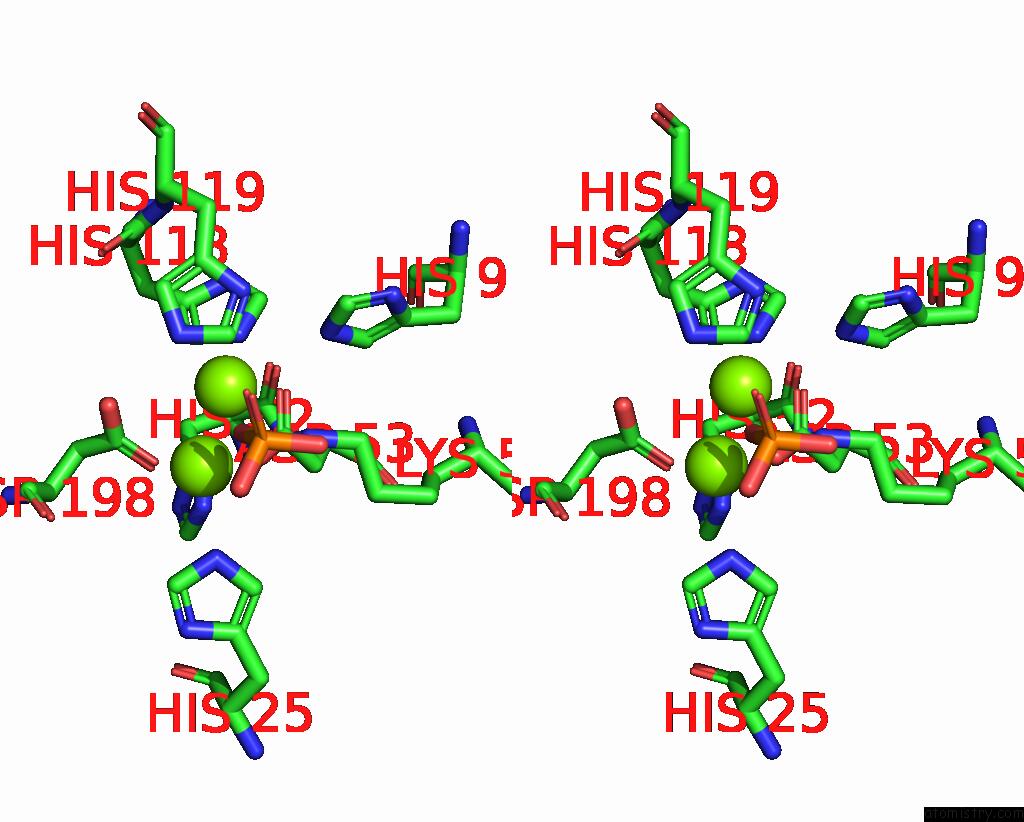
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 1 of Exploiting Activation and Inactivation Mechanisms in Type I-C Crispr- CAS3 For Genome Editing Applications within 5.0Å range:
|
Magnesium binding site 2 out of 2 in 8g9u
Go back to
Magnesium binding site 2 out
of 2 in the Exploiting Activation and Inactivation Mechanisms in Type I-C Crispr- CAS3 For Genome Editing Applications
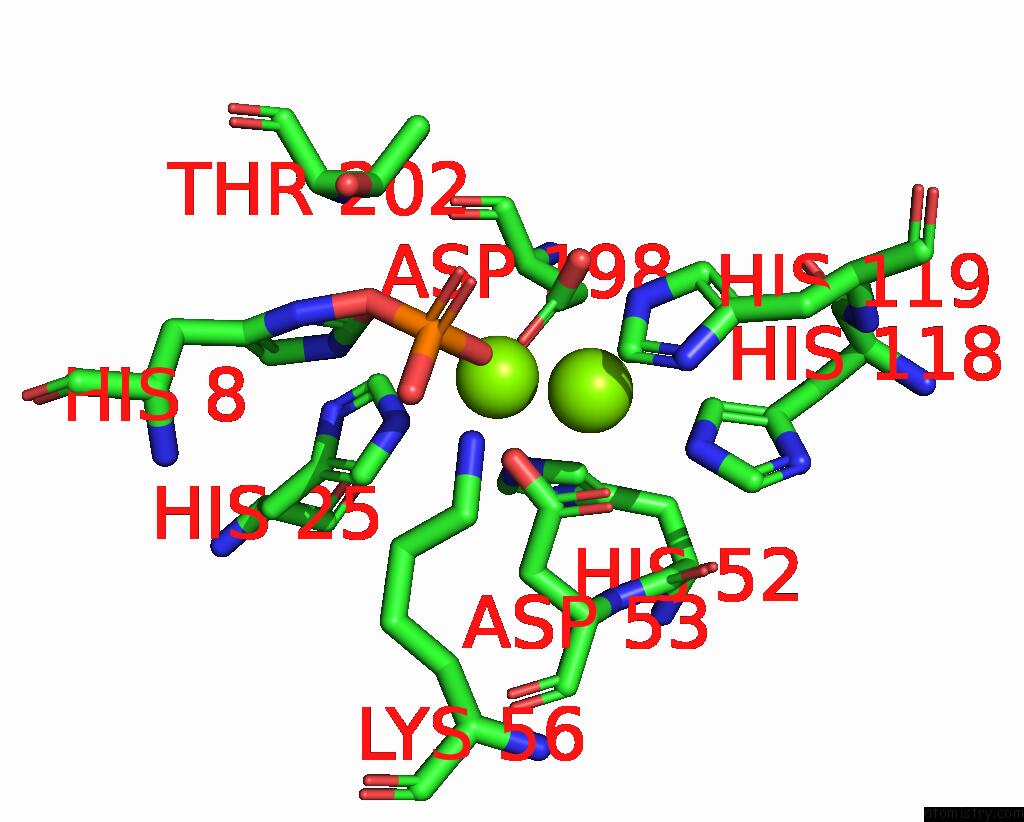
Mono view
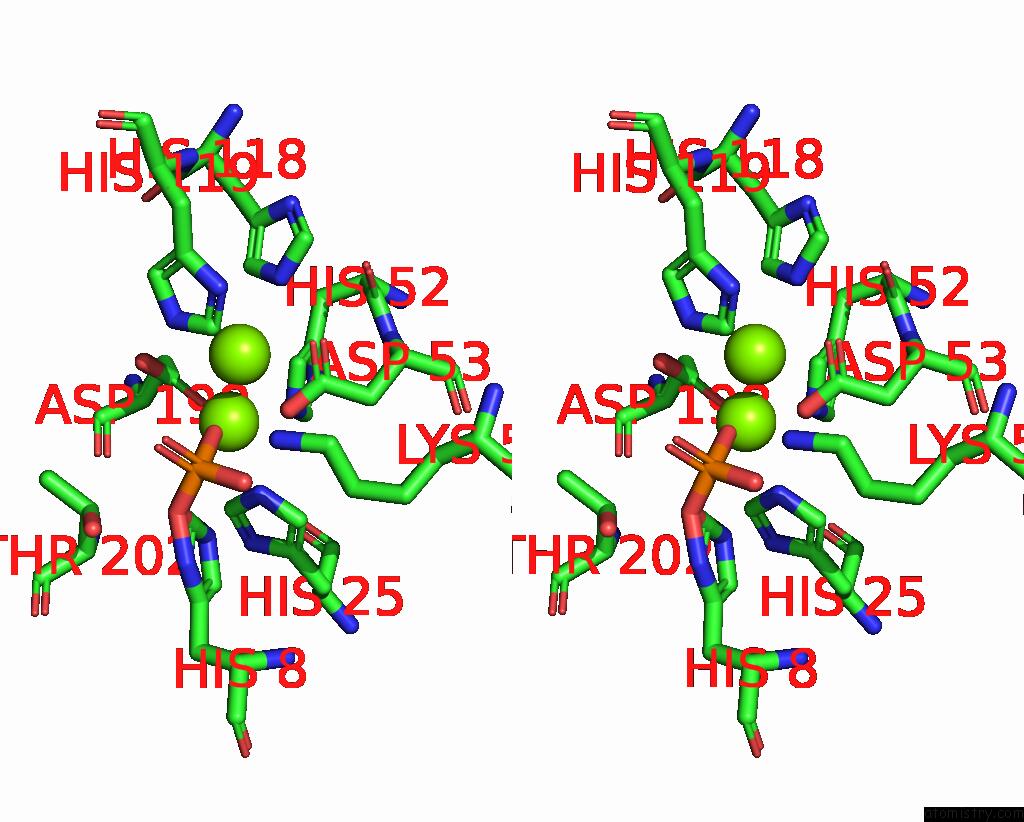
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 2 of Exploiting Activation and Inactivation Mechanisms in Type I-C Crispr- CAS3 For Genome Editing Applications within 5.0Å range:
|
Reference:
C.Hu,
M.T.Myers,
X.Zhou,
Z.Hou,
M.L.Lozen,
K.H.Nam,
Y.Zhang,
A.Ke.
Exploiting Activation and Inactivation Mechanisms in Type I-C Crispr-CAS3 For Genome-Editing Applications. Mol.Cell V. 84 463 2024.
ISSN: ISSN 1097-2765
PubMed: 38242128
DOI: 10.1016/J.MOLCEL.2023.12.034
Page generated: Fri Oct 4 03:06:09 2024
ISSN: ISSN 1097-2765
PubMed: 38242128
DOI: 10.1016/J.MOLCEL.2023.12.034
Last articles
Mg in 5JWBMg in 5JWA
Mg in 5JVL
Mg in 5JVN
Mg in 5JVM
Mg in 5JVJ
Mg in 5JVD
Mg in 5JV5
Mg in 5JTG
Mg in 5JRW