Magnesium »
PDB 8p1m-8p5w »
8p2i »
Magnesium in PDB 8p2i: Cryo-Em Structure of Pyrococcus Furiosus Apo Form Rna Polymerase Contracted Clamp Conformation with SPT4/5
Enzymatic activity of Cryo-Em Structure of Pyrococcus Furiosus Apo Form Rna Polymerase Contracted Clamp Conformation with SPT4/5
All present enzymatic activity of Cryo-Em Structure of Pyrococcus Furiosus Apo Form Rna Polymerase Contracted Clamp Conformation with SPT4/5:
2.7.7.6;
2.7.7.6;
Other elements in 8p2i:
The structure of Cryo-Em Structure of Pyrococcus Furiosus Apo Form Rna Polymerase Contracted Clamp Conformation with SPT4/5 also contains other interesting chemical elements:
| Zinc | (Zn) | 6 atoms |
Magnesium Binding Sites:
The binding sites of Magnesium atom in the Cryo-Em Structure of Pyrococcus Furiosus Apo Form Rna Polymerase Contracted Clamp Conformation with SPT4/5
(pdb code 8p2i). This binding sites where shown within
5.0 Angstroms radius around Magnesium atom.
In total only one binding site of Magnesium was determined in the Cryo-Em Structure of Pyrococcus Furiosus Apo Form Rna Polymerase Contracted Clamp Conformation with SPT4/5, PDB code: 8p2i:
In total only one binding site of Magnesium was determined in the Cryo-Em Structure of Pyrococcus Furiosus Apo Form Rna Polymerase Contracted Clamp Conformation with SPT4/5, PDB code: 8p2i:
Magnesium binding site 1 out of 1 in 8p2i
Go back to
Magnesium binding site 1 out
of 1 in the Cryo-Em Structure of Pyrococcus Furiosus Apo Form Rna Polymerase Contracted Clamp Conformation with SPT4/5
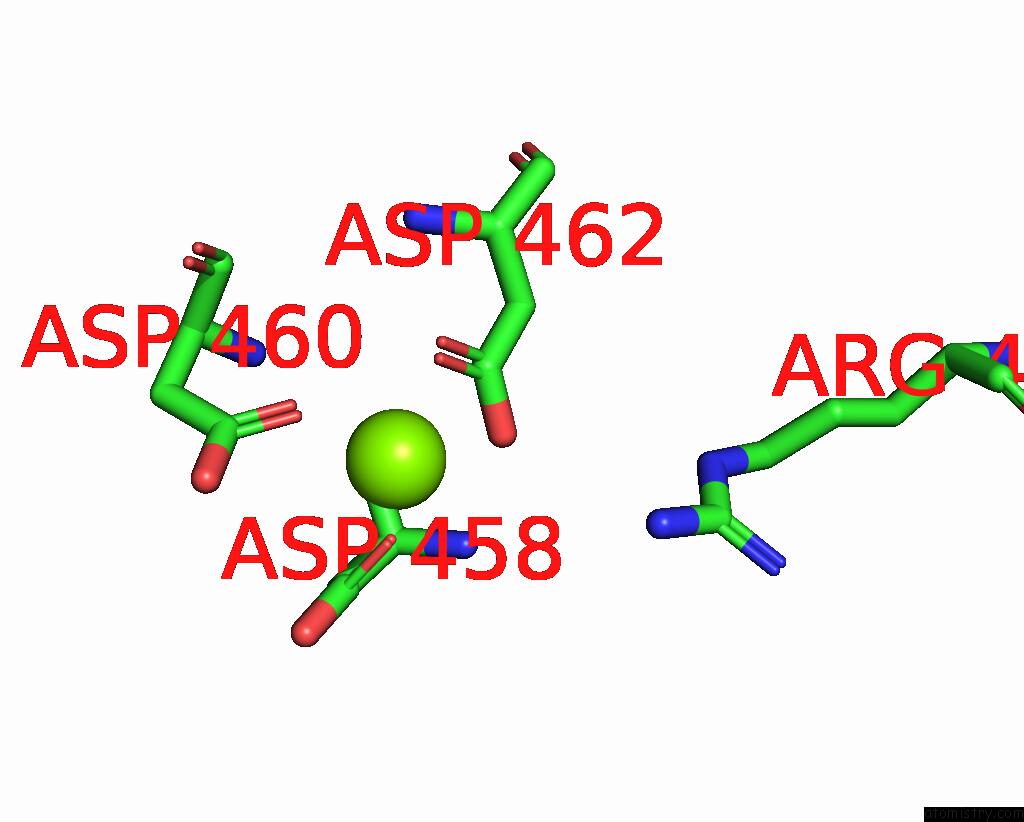
Mono view
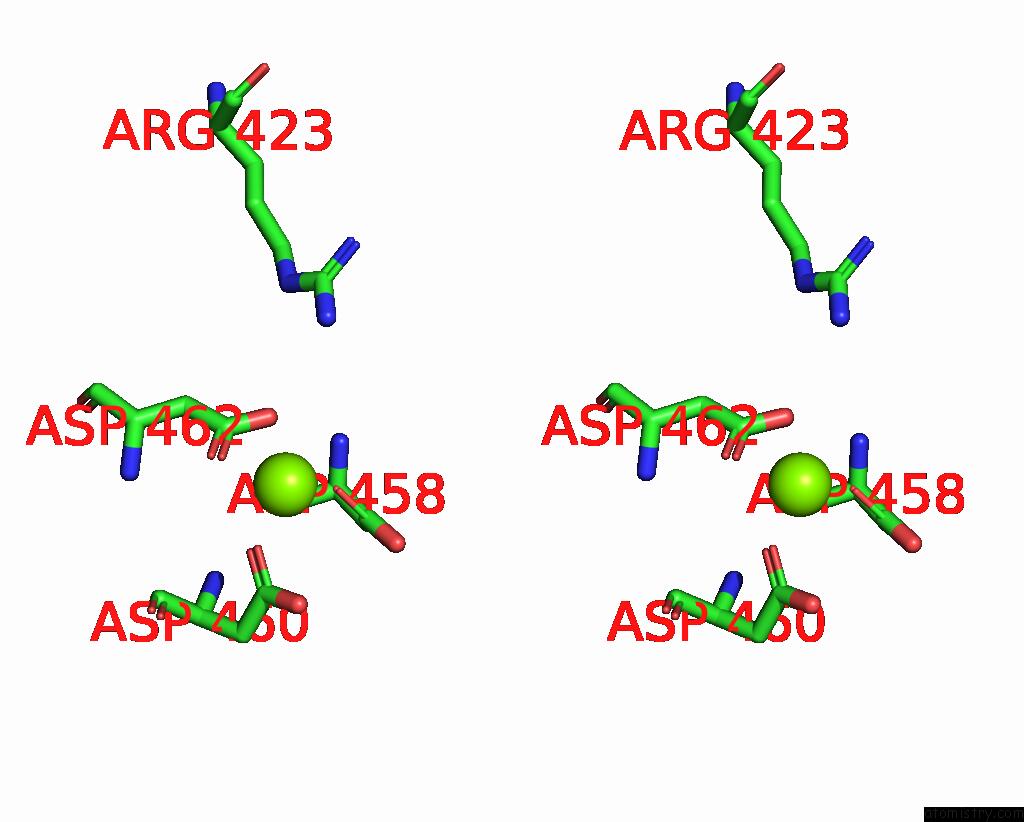
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 1 of Cryo-Em Structure of Pyrococcus Furiosus Apo Form Rna Polymerase Contracted Clamp Conformation with SPT4/5 within 5.0Å range:
|
Reference:
D.M.Tarau,
F.Grunberger,
M.Pilsl,
R.Reichelt,
F.B.Heiss,
S.Koenig,
H.Urlaub,
W.Hausner,
C.Engel,
D.Grohmann.
Structural Basis of Archaeal Rna Polymerase Transcription Elongation and SPT4/5 Recruitment Nucleic Acids Res. 2024.
ISSN: ESSN 1362-4962
Page generated: Fri Aug 15 12:06:12 2025
ISSN: ESSN 1362-4962
Last articles
Mn in 9LJUMn in 9LJW
Mn in 9LJS
Mn in 9LJR
Mn in 9LJT
Mn in 9LJV
Mg in 9UA2
Mg in 9R96
Mg in 9VM1
Mg in 9P01