Magnesium »
PDB 8syi-8t7p »
8t02 »
Magnesium in PDB 8t02: Reconstituted E. Coli Rna Polymerase Post-Termination Complex on Negatively-Supercoiled Dna: Unwinding Duplex Dna (Rptci)
Enzymatic activity of Reconstituted E. Coli Rna Polymerase Post-Termination Complex on Negatively-Supercoiled Dna: Unwinding Duplex Dna (Rptci)
All present enzymatic activity of Reconstituted E. Coli Rna Polymerase Post-Termination Complex on Negatively-Supercoiled Dna: Unwinding Duplex Dna (Rptci):
2.7.7.6;
2.7.7.6;
Other elements in 8t02:
The structure of Reconstituted E. Coli Rna Polymerase Post-Termination Complex on Negatively-Supercoiled Dna: Unwinding Duplex Dna (Rptci) also contains other interesting chemical elements:
| Zinc | (Zn) | 2 atoms |
Magnesium Binding Sites:
The binding sites of Magnesium atom in the Reconstituted E. Coli Rna Polymerase Post-Termination Complex on Negatively-Supercoiled Dna: Unwinding Duplex Dna (Rptci)
(pdb code 8t02). This binding sites where shown within
5.0 Angstroms radius around Magnesium atom.
In total only one binding site of Magnesium was determined in the Reconstituted E. Coli Rna Polymerase Post-Termination Complex on Negatively-Supercoiled Dna: Unwinding Duplex Dna (Rptci), PDB code: 8t02:
In total only one binding site of Magnesium was determined in the Reconstituted E. Coli Rna Polymerase Post-Termination Complex on Negatively-Supercoiled Dna: Unwinding Duplex Dna (Rptci), PDB code: 8t02:
Magnesium binding site 1 out of 1 in 8t02
Go back to
Magnesium binding site 1 out
of 1 in the Reconstituted E. Coli Rna Polymerase Post-Termination Complex on Negatively-Supercoiled Dna: Unwinding Duplex Dna (Rptci)
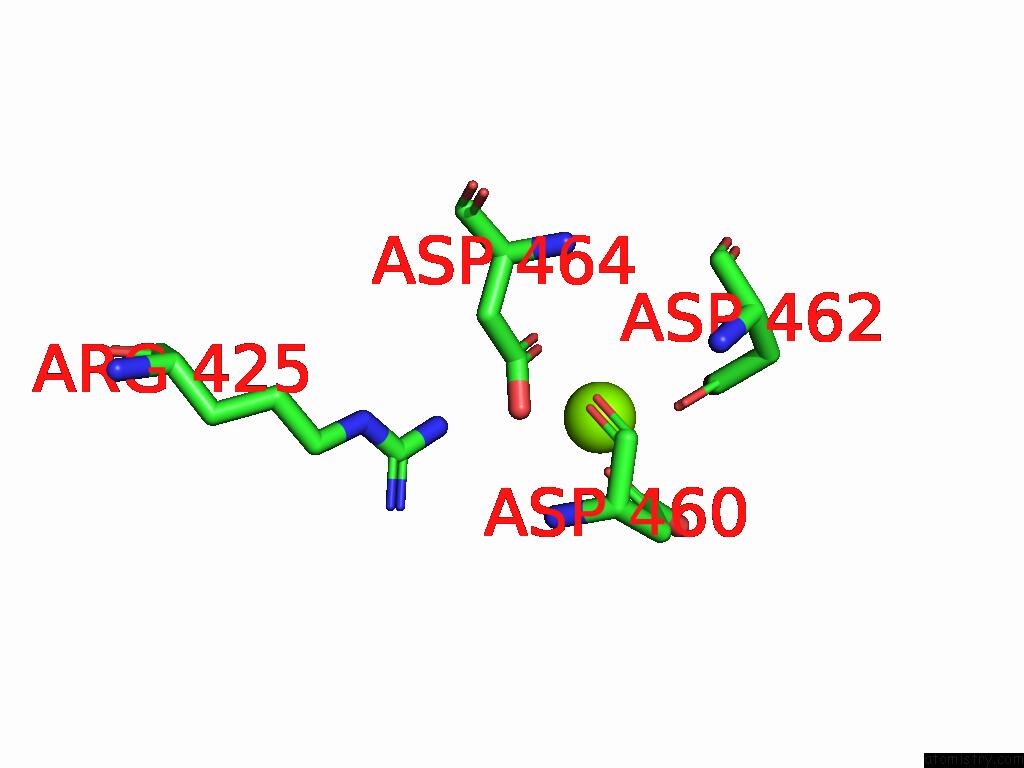
Mono view
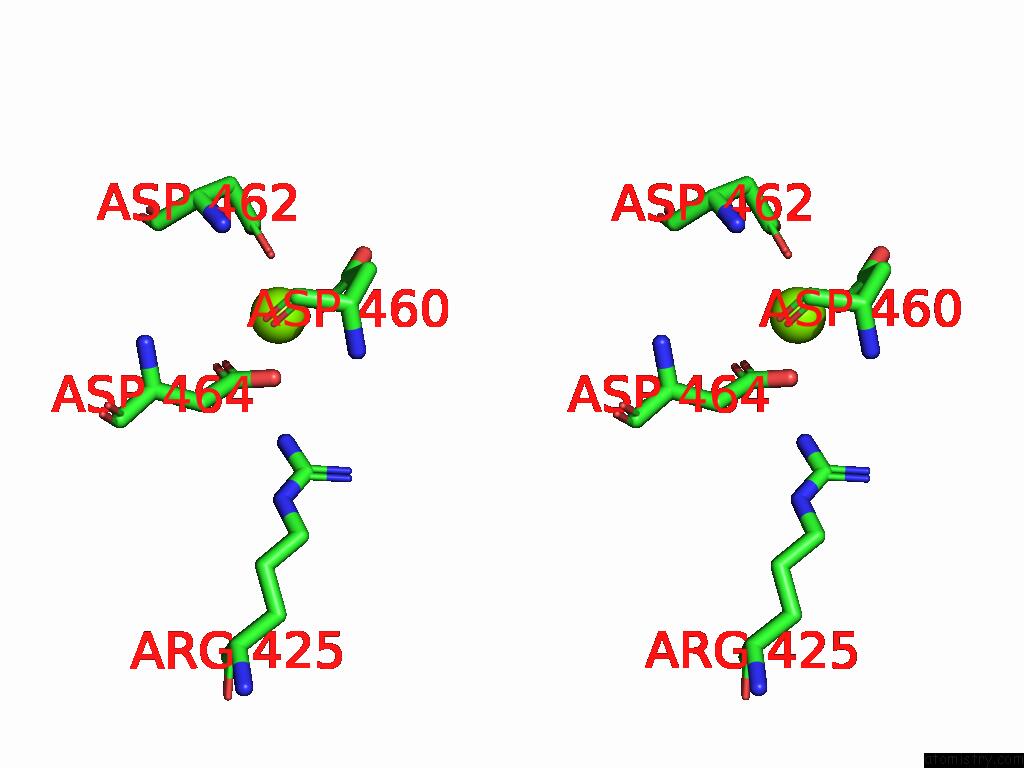
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 1 of Reconstituted E. Coli Rna Polymerase Post-Termination Complex on Negatively-Supercoiled Dna: Unwinding Duplex Dna (Rptci) within 5.0Å range:
|
Reference:
J.J.Brewer,
K.Inlow,
R.A.Mooney,
B.Bosch,
P.D.B.Olinares,
L.P.Marcelino,
B.T.Chait,
R.Landick,
J.Gelles,
E.A.Campbell,
S.A.Darst.
Rapa Opens the Rna Polymerase Clamp to Disrupt Post-Termination Complexes and Prevent Cytotoxic R-Loop Formation. Nat.Struct.Mol.Biol. 2025.
ISSN: ESSN 1545-9985
PubMed: 39779919
DOI: 10.1038/S41594-024-01447-8
Page generated: Fri Aug 15 16:04:26 2025
ISSN: ESSN 1545-9985
PubMed: 39779919
DOI: 10.1038/S41594-024-01447-8
Last articles
Mn in 2DFJMn in 2DTI
Mn in 2D3C
Mn in 2DF4
Mn in 2D3A
Mn in 2DEG
Mn in 2D3B
Mn in 2D7R
Mn in 2D7I
Mn in 2D7F